EQUIX: Jq divergence#
Show code cell content
%load_ext watermark
%matplotlib inline
import glob
import os
import cf_xarray as cfxr
import dcpy
import distributed
import hvplot.xarray
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
from dcpy.oceans import read_osu_microstructure_mat
from IPython.display import Image
# import eddydiff as ed
import pump
xr.set_options(keep_attrs=True)
mpl.rcParams["figure.dpi"] = 140
%watermark -iv
The watermark extension is already loaded. To reload it, use:
%reload_ext watermark
xarray : 0.19.0
cf_xarray : 0.5.3.dev29+g3660810.d20210729
numpy : 1.21.3
hvplot : 0.7.3
dcpy : 0.1
matplotlib : 3.4.3
distributed: 2021.10.0
pump : 0.1
Read data#
Show code cell content
# th = xr.load_dataset("/home/deepak/datasets/microstructure/osu/tropicheat.nc")
# tiwe = xr.load_dataset("/home/deepak/datasets/microstructure/osu/tiwe.nc")
equix = xr.load_dataset("/home/deepak/datasets/microstructure/osu/equix.nc")
tao = xr.open_dataset(
"/home/deepak/work/datasets/microstructure/osu/equix/hourly_tao.nc"
)
iop = xr.open_dataset(
"/home/deepak/work/datasets/microstructure/osu/equix/hourly_iop.nc"
)
eop = xr.open_dataset(
"/home/deepak/work/datasets/microstructure/osu/equix/hourly_eop.nc"
)
%matplotlib inline
plt.rcParams["figure.dpi"] = 140
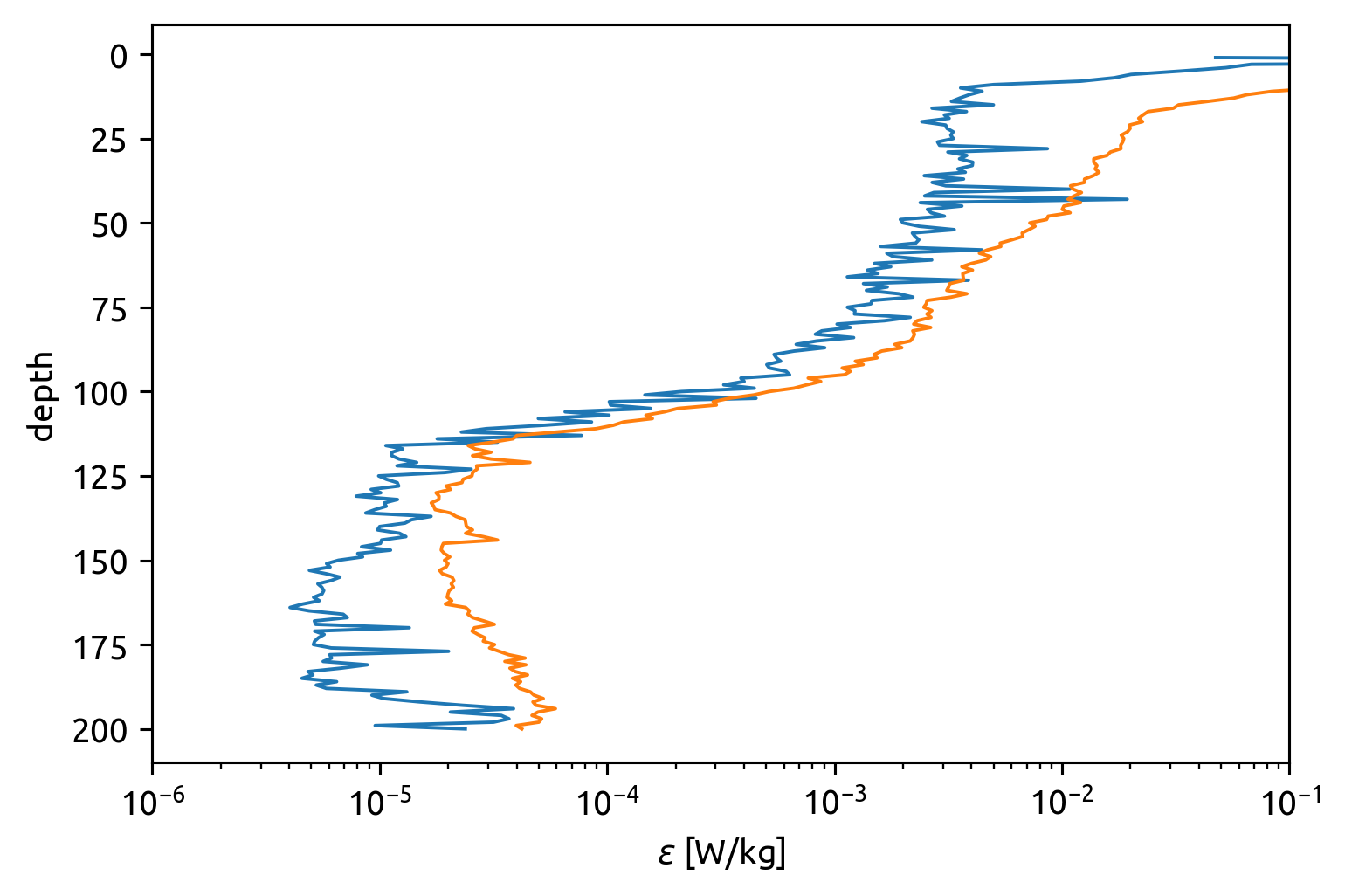
np.abs(equix.chi / 2 / (equix.dTdz.where(np.abs(equix.dTdz) > 5e-3) ** 2)).mean(
"time"
).plot(y="depth", yincrease=False)
np.abs(0.2 * equix.eps / equix.N2).mean("time").plot(
y="depth", yincrease=False, xscale="log", xlim=(1e-6, 1e-1)
)
[<matplotlib.lines.Line2D at 0x7f8ecf07cd90>]
Chameleon dJ/dz estimate#
subset = equix.sel(time=slice("2008-10-29", "2008-11-03"))
eps_plot = subset.eps.hvplot.quadmesh(
y="depth",
x="time",
cmap="RdYlBu_r",
cnorm="log",
clim=(1e-10, 1e-5),
ylim=(200, 0),
)
theta_contours = subset.theta.hvplot.contour(
# levels=[21, 22, 23, 24], #colors="gray", #linewidths=0.5, x="time", ax=ax[0]
)
# equix.theta.cf.plot.contour(
# levels=bins, colors="k", linewidths=0.25, x="time", ax=ax[0]
# )
# equix.mld.plot(color="g", ax=ax[0], lw=0.25)
# equix.eucmax.plot(color="k", ax=ax[0], lw=0.5)
# dcpy.plots.liney(iop.depth.data, zorder=10, color="w", ax=ax[0])
# dcpy.plots.liney(tao.depth.data, zorder=10, color="cyan", ax=ax[0])
eps_plot * theta_contours