Heat and momentum budgets with MITgcm output#
Long names for all variables are in available_diagnostics.log
imports and cluster setup#
%load_ext autoreload
%autoreload 2
%matplotlib inline
import dask
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import seawater as sw
import xarray as xr
# import hvplot.xarray
import dcpy
import pump
# import facetgrid
mpl.rcParams["savefig.dpi"] = 300
mpl.rcParams["savefig.bbox"] = "tight"
mpl.rcParams["figure.dpi"] = 120
xr.set_options(keep_attrs=False)
import distributed
import dask_jobqueue
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
if "client" in locals():
client.close()
cluster.close()
env = {"OMP_NUM_THREADS": "3", "NUMBA_NUM_THREADS": "3"}
cluster = distributed.LocalCluster(n_workers=12, threads_per_worker=1, env=env)
# cluster = dask_jobqueue.SLURMCluster(
# cores=1, processes=1, memory="25GB", walltime="02:00:00", project="NCGD0011"
# )
# cluster = dask_jobqueue.PBSCluster(
# cores=9, processes=9, memory="108GB", walltime="02:00:00", project="NCGD0043",
# env_extra=env,
# )
client = distributed.Client(cluster)
cluster
Utility functions#
read_metrics will sometimes fail because the files don’t exist. This happens when Scott restarts a run :)
def read_metrics(dirname, longitude, latitude, depth):
"""
This function needs longitude, latitude, depth to assign the right metadata.
If size of the metrics variables are not the same as (longitude, latitude),
the code assumes that a boundary region has been cut out at the low-end and
high-end of the appropriate axis.
If the size in depth-axis is different, then it assumes that the provided depth
is a slice from surface to the Nth-point where N=len(depth).
"""
import xmitgcm
h = dict()
for ff in ["hFacC", "RAC", "RF", "DXC", "DYC"]:
try:
h[ff] = xmitgcm.utils.read_mds(dirname + ff)[ff]
except FileNotFoundError:
print(f"metrics files not available. {dirname + ff}")
metrics = None
return xr.Dataset()
hFacC = h["hFacC"].copy().squeeze().astype("float32")
RAC = h["RAC"].copy().squeeze().astype("float32")
RF = h["RF"].copy().squeeze().astype("float32")
DXC = h["DXC"].copy().squeeze().astype("float32")
DYC = h["DYC"].copy().squeeze().astype("float32")
del h
if len(longitude) != RAC.shape[1]:
dlon = RAC.shape[1] - len(longitude)
lons = slice(dlon // 2, -dlon // 2)
else:
lons = slice(None, None)
if len(latitude) != RAC.shape[0]:
dlat = RAC.shape[0] - len(latitude)
lats = slice(dlat // 2, -dlat // 2)
else:
lats = slice(None, None)
RAC = xr.DataArray(
RAC[lats, lons],
dims=["latitude", "longitude"],
coords={"longitude": longitude, "latitude": latitude},
name="RAC",
)
DXC = xr.DataArray(
DXC[lats, lons],
dims=["latitude", "longitude"],
coords={"longitude": longitude, "latitude": latitude},
name="DXC",
)
DYC = xr.DataArray(
DYC[lats, lons],
dims=["latitude", "longitude"],
coords={"longitude": longitude, "latitude": latitude},
name="DYC",
)
depth = xr.DataArray(
(RF[1:] + RF[:-1]) / 2,
dims=["depth"],
name="depth",
attrs={"long_name": "depth", "units": "m"},
)
dRF = xr.DataArray(
np.diff(RF.squeeze()),
dims=["depth"],
coords={"depth": depth},
name="dRF",
attrs={"long_name": "cell_height", "units": "m"},
)
RF = xr.DataArray(RF.squeeze(), dims=["depth_left"], name="depth_left")
hFacC = xr.DataArray(
hFacC[:, lats, lons],
dims=["depth", "latitude", "longitude"],
coords={
"depth": depth,
"latitude": latitude,
"longitude": longitude,
},
name="hFacC",
)
metrics = xr.merge([dRF, hFacC, RAC, DXC, DYC])
metrics["cellvol"] = np.abs(metrics.RAC * metrics.dRF * metrics.hFacC)
metrics["cellvol"] = metrics.cellvol.where(metrics.cellvol > 0)
metrics["RF"] = RF
metrics["rAw"] = xr.DataArray(
xmitgcm.utils.read_mds(dirname + "/RAW")["RAW"][lats, lons].astype("float32"),
dims=["latitude", "longitude"],
)
metrics["hFacW"] = xr.DataArray(
xmitgcm.utils.read_mds(dirname + "/hFacW")["hFacW"][:, lats, lons].astype(
"float32"
),
dims=["depth", "latitude", "longitude"],
)
metrics["hFacW"] = metrics.hFacW.where(metrics.hFacW > 0)
metrics["drF"] = xr.DataArray(
xmitgcm.utils.read_mds(dirname + "/DRF")["DRF"].squeeze().astype("float32"),
dims=["depth"],
)
metrics = metrics.isel(
depth=slice(budget.sizes["depth"]), depth_left=slice(budget.sizes["depth"] + 1)
)
return metrics
def plot_lhs_rhs(LHS, RHS):
LHS, RHS = dask.compute(LHS, RHS)
kwargs = dict(histtype="step", bins=1000, density=True, ylim=[0, 5])
f, ax = plt.subplots(2, 1, constrained_layout=True)
np.log10(np.abs(LHS).where(np.abs(LHS) > 0)).plot.hist(ax=ax[0], **kwargs)
np.log10(np.abs(RHS).where(np.abs(RHS) > 0)).plot.hist(ax=ax[0], **kwargs)
diff = np.abs(LHS - RHS)
np.log10(diff.where(diff > 0)).plot.hist(ax=ax[0], **kwargs)
ax[0].legend(("LHS", "RHS", "LHS-RHS"))
if diff.ndim == 2:
(LHS - RHS).plot(ax=ax[1], x="longitude", robust=True)
ax[1].set_title("LHS - RHS")
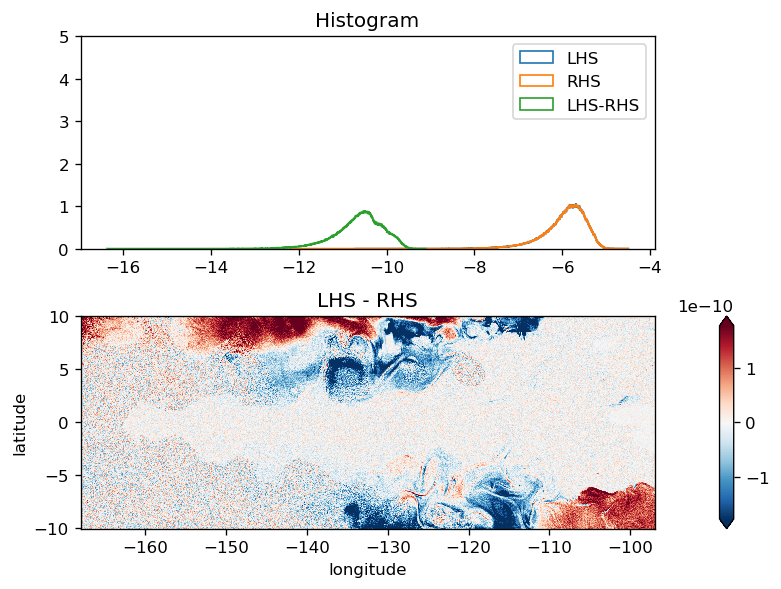
Heat budget#
dirname = "/glade/campaign/cgd/oce/people/bachman/TPOS_MITgcm_fix2/"
Read in data#
hb_files = xr.open_dataset(
dirname + "File_0003_hb.nc", chunks={"latitude": 120, "longitude": 500}
)
sf_files = xr.open_dataset(
dirname + "File_0003_surf.nc", chunks={"latitude": 120, "longitude": 500}
)
budget = xr.merge([hb_files, sf_files]).transpose(
"longitude", "latitude", "depth", "time"
)
budget["oceQsw"] = budget.oceQsw.fillna(0)
# budget = budget.drop(["DFxE_TH", "DFyE_TH", "DFrE_TH"])
metrics = read_metrics(
dirname,
longitude=budget.longitude,
latitude=budget.latitude,
depth=budget.depth,
).chunk({"latitude": 120, "longitude": 500})
def sw_prof(depth):
"""MITgcm Shortwave radiation penetration profile."""
return 0.62 * np.exp(depth / 0.6) + (1 - 0.62) * np.exp(depth / 20)
# penetrative shortwave radiation
budget["swprofile"] = xr.DataArray(
sw_prof(metrics.RF[:-1].values) - sw_prof(metrics.RF[1:].values),
dims=["depth"],
attrs={"long_name": "SW radiation deposited in cell"},
)
budget
<xarray.Dataset>
Dimensions: (depth: 136, latitude: 400, longitude: 1420, time: 1)
Coordinates:
* depth (depth) float64 -1.25 -3.75 -6.25 -8.75 ... -824.4 -881.7 -944.4
* latitude (latitude) float32 -10.0 -9.949875 -9.89975 ... 9.949875 10.0
* longitude (longitude) float32 -168.0 -167.94997 ... -97.05003 -97.0
* time (time) datetime64[ns] 1999-01-03
Data variables:
TOTTTEND (longitude, latitude, depth, time) float32 dask.array<chunksize=(500, 120, 136, 1), meta=np.ndarray>
ADVx_TH (longitude, latitude, depth, time) float32 dask.array<chunksize=(500, 120, 136, 1), meta=np.ndarray>
ADVy_TH (longitude, latitude, depth, time) float32 dask.array<chunksize=(500, 120, 136, 1), meta=np.ndarray>
ADVr_TH (longitude, latitude, depth, time) float32 dask.array<chunksize=(500, 120, 136, 1), meta=np.ndarray>
DFrI_TH (longitude, latitude, depth, time) float32 dask.array<chunksize=(500, 120, 136, 1), meta=np.ndarray>
KPPg_TH (longitude, latitude, depth, time) float32 dask.array<chunksize=(500, 120, 136, 1), meta=np.ndarray>
WTHMASS (longitude, latitude, depth, time) float32 dask.array<chunksize=(500, 120, 136, 1), meta=np.ndarray>
oceQsw (longitude, latitude, time) float32 dask.array<chunksize=(500, 120, 1), meta=np.ndarray>
TFLUX (longitude, latitude, time) float32 dask.array<chunksize=(500, 120, 1), meta=np.ndarray>
swprofile (depth) float32 0.65503883 0.04886788 ... 5.854533e-21test balances#
The heat budget code is dirtier than the momentum budget and could be cleanedup a bit
level = 120
levelp = level + 1
terms = budget.isel(depth=level).squeeze()
metrics_sub = metrics.isel(depth=level, latitude=slice(1, -1), longitude=slice(1, -1))
cellvol = metrics_sub.cellvol.transpose()
terms["ADVr_THp"] = budget["ADVr_TH"].isel(depth=levelp).squeeze()
terms["DFrI_THp"] = budget["DFrI_TH"].isel(depth=levelp).squeeze()
terms["KPPg_THp"] = budget["KPPg_TH"].isel(depth=levelp).squeeze()
surf_mass = terms.WTHMASS[1:-1, 1:-1] * metrics_sub.RAC[1:-1, 1:-1]
# constants taken from diagnostics.log (I think)
global_area = 2.196468634481708e13
rhoConst = 1035
Cp = 3994
TsurfCor = surf_mass.sum() / global_area
LHS = terms.TOTTTEND[1:-1, 1:-1] / 86400
ADVx = (terms.ADVx_TH.values[2:, 1:-1] - terms.ADVx_TH.values[1:-1, 1:-1]) / cellvol
ADVy = (terms.ADVy_TH.values[1:-1, 2:] - terms.ADVy_TH.values[1:-1, 1:-1]) / cellvol
ADVr = (
terms.ADVr_TH.fillna(0).values[1:-1, 1:-1] - terms.ADVr_THp.values[1:-1, 1:-1]
) / cellvol
# no explicit diffusion
# DFxE = (terms.DFxE_TH.values[2:, 1:-1] - terms.DFxE_TH.values[1:-1, 1:-1]) / cellvol
# DFyE = (terms.DFyE_TH.values[1:-1, 2:] - terms.DFyE_TH.values[1:-1, 1:-1]) / cellvol
# DFrE = (terms.DFrE_TH.values[1:-1, 1:-1] - terms.DFrE_THp.values[1:-1, 1:-1]) / cellvol
DFrI = (
terms.DFrI_TH.fillna(0).values[1:-1, 1:-1] - terms.DFrI_THp.values[1:-1, 1:-1]
) / cellvol
# non-local stuff only in mixing layer
KPPg = (
terms.KPPg_TH.fillna(0).values[1:-1, 1:-1]
- terms.KPPg_THp.fillna(0).values[1:-1, 1:-1]
) / cellvol
ADV = LHS.copy(data=ADVx + ADVy + ADVr)
DIFF = LHS.copy(data=DFrI + KPPg) # no explicit horizontal diffusion
ADV.name = "ADV"
DIFF.name = "DIFF"
SW = (
terms.oceQsw[1:-1, 1:-1]
/ (rhoConst * Cp)
/ (metrics_sub.dRF * metrics_sub.hFacC)
* terms.swprofile
)
RHS = ADV + DIFF + SW
# surface level stuff
if level == 0:
# surface tendency due to SW flux
tflx_tend = (terms.TFLUX[1:-1, 1:-1] - terms.oceQsw[1:-1, 1:-1]) / (
rhoConst * Cp * metrics_sub.dRF * metrics_sub.hFacC
)
# tendency due to mass correction
surf_corr_tend = -(
(terms.WTHMASS[1:-1, 1:-1] - TsurfCor) / (metrics_sub.dRF * metrics_sub.hFacC)
)
RHS += tflx_tend + surf_corr_tend
RHS *= -1
plot_lhs_rhs(LHS, RHS)
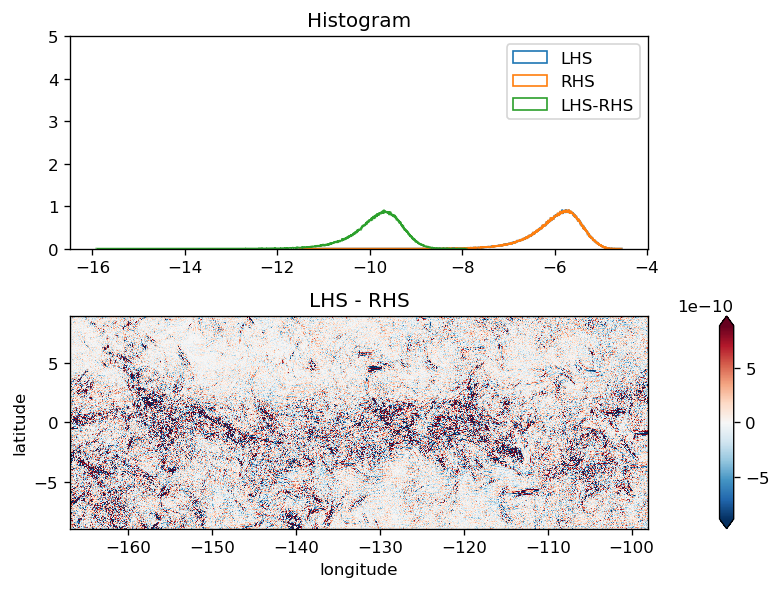
Momentum budget#
Momentum budget emails:
http://mailman.mitgcm.org/pipermail/mitgcm-support/2010-December/006921.html
http://mailman.mitgcm.org/pipermail/mitgcm-support/2013-December/008702.html
http://mailman.mitgcm.org/pipermail/mitgcm-support/2017-July/011196.html
http://mailman.mitgcm.org/pipermail/mitgcm-devel/2019-June/007077.html
read in data#
Need to append zeros to the viscosity terms because the bottom level is not stored :/
ssh_files = xr.open_mfdataset(
dirname + "File_*_etan.nc",
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
)
ub_files = xr.open_mfdataset(
dirname + "File_*_ub.nc",
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
)
vb_files = xr.open_mfdataset(
dirname + "File_*_vb.nc",
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
)
# sf_files = xr.open_mfdataset(dirname + 'File_*_surf.nc', chunks={"latitude": 120, "longitude": 500}, combine="by_coords", parallel=True)[["ETAN"]]
budget = (
xr.merge([ub_files, vb_files, ssh_files]).transpose(
"longitude", "latitude", "depth", "time"
)
).squeeze()
# From http://mailman.mitgcm.org/pipermail/mitgcm-support/2010-December/006920.html
# And to get a tendency from this vertical flux:
# Um_Impl(:,:,k) =[ VISrI_Um(:,:,k+1) - VISrI_Um(:,:,k) ]
# /[ rAw(:,:)*drF(k)*hFacW(:,:,k) ]
# (hope I got the sign right)
# VISrI_Um(k=1) should be zero, and the bottom one (k=Nr+1)
# is not shored in the output file but it's also zero.
budget["VISrI_Um"] = budget.VISrI_Um.fillna(0) # fills surface values with 0
budget["VISrI_Vm"] = budget.VISrI_Vm.fillna(0)
# NOTE: This is totally wrong now since TPOS_MITgcm_20 is only a top 1000m subset.
VISrI_Um = xr.concat(
[
budget.VISrI_Um,
xr.zeros_like(budget.VISrI_Um.isel(depth=0).drop("depth")).expand_dims(
depth=[-5865.922]
),
],
dim="depth",
)
VISrI_Vm = xr.concat(
[
budget.VISrI_Vm,
xr.zeros_like(budget.VISrI_Vm.isel(depth=0).drop("depth")).expand_dims(
depth=[-5865.922]
),
],
dim="depth",
)
metrics = read_metrics(
dirname, longitude=budget.longitude, latitude=budget.latitude, depth=budget.depth
).chunk({"latitude": 120, "longitude": 500})
metrics
<xarray.Dataset>
Dimensions: (depth: 136, depth_left: 137, latitude: 400, longitude: 1420)
Coordinates:
* depth (depth) float64 -1.25 -3.75 -6.25 -8.75 ... -824.4 -881.7 -944.4
* latitude (latitude) float64 -10.0 -9.95 -9.9 -9.85 ... 9.85 9.9 9.95 10.0
* longitude (longitude) float64 -168.0 -167.9 -167.9 ... -97.1 -97.05 -97.0
Dimensions without coordinates: depth_left
Data variables:
dRF (depth) float32 dask.array<chunksize=(136,), meta=np.ndarray>
hFacC (depth, latitude, longitude) float32 dask.array<chunksize=(136, 120, 500), meta=np.ndarray>
RAC (latitude, longitude) float32 dask.array<chunksize=(120, 500), meta=np.ndarray>
DXC (latitude, longitude) float32 dask.array<chunksize=(120, 500), meta=np.ndarray>
DYC (latitude, longitude) float32 dask.array<chunksize=(120, 500), meta=np.ndarray>
cellvol (latitude, longitude, depth) float32 dask.array<chunksize=(120, 500, 136), meta=np.ndarray>
RF (depth_left) float32 dask.array<chunksize=(137,), meta=np.ndarray>
rAw (latitude, longitude) float32 dask.array<chunksize=(120, 500), meta=np.ndarray>
hFacW (depth, latitude, longitude) float32 dask.array<chunksize=(136, 120, 500), meta=np.ndarray>
drF (depth) float32 dask.array<chunksize=(136,), meta=np.ndarray>U momentum#
We are using implicSurfPres=1 and Scott is saving SSH at the appropriate timestep.
level = 120
# depth level # budget.sizes["depth"]
# SSHx = - 9.81 * budget.ETAN.copy(
# data=np.gradient(budget.ETAN, axis=budget.ETAN.get_axis_num("longitude"))
# ) / metrics.DXC
# when trying to do a budget over just 1 time-step, one needs to be aware
# of the precise time-discretisation of the the first term which is evaluated
# in the model as
# - gravity * { implicSurfPress * (d.EtaN/dx)^n+1
# +(1-implicSurfPress)*(d.EtaN/dx)^n }
# (using the default implicSurfPress=1 , it might be be easier to use
# snap-shot output Eta.[iter+1] to get EtaN^n+1 )
# this works because Scott has saved SSH one timestep prior to the saved timestep for the other fields
SSHx = (
-9.81
* budget.ETAN.diff("longitude").reindex(longitude=budget.longitude)
/ metrics.DXC
)
# from http://mailman.mitgcm.org/pipermail/mitgcm-support/2010-December/006921.html
Um_Impl = VISrI_Um.diff("depth", label="lower") / (
metrics.rAw * metrics.drF.isel(depth=level) * metrics.hFacW.isel(depth=level)
)
RHS = (
SSHx
+ budget.Um_dPHdx
+ budget.Um_Advec
# + budget.Um_Cori # is in Um_Advec
+ budget.Um_Diss
+ Um_Impl
+ budget.Um_Ext.fillna(0)
+ budget.AB_gU
).isel(depth=level)
LHS = budget.TOTUTEND.isel(depth=level) / 86400
# da = xr.Dataset({"LHS": LHS, "RHS": RHS}).to_array("variable")
# da.compute().plot(row="variable", robust=True, x="longitude", cbar_kwargs={"orientation": "horizontal"})
/glade/u/home/dcherian/miniconda3/envs/dcpy/lib/python3.6/site-packages/dask/array/core.py:3908: PerformanceWarning: Increasing number of chunks by factor of 20
**blockwise_kwargs,
(LHS - RHS).isel(time=slice(12)).compute().plot(
col="time",
col_wrap=4,
cbar_kwargs={"orientation": "horizontal"},
x="longitude",
robust=True,
)
Task was destroyed but it is pending!
task: <Task pending coro=<Nanny._on_exit() running at /glade/u/home/dcherian/miniconda3/envs/dcpy/lib/python3.6/site-packages/distributed/nanny.py:406> wait_for=<Future pending cb=[<TaskWakeupMethWrapper object at 0x2b8d2c255918>()]> cb=[IOLoop.add_future.<locals>.<lambda>() at /glade/u/home/dcherian/miniconda3/envs/dcpy/lib/python3.6/site-packages/tornado/ioloop.py:690]>
<xarray.plot.facetgrid.FacetGrid at 0x2b8db53452e8>
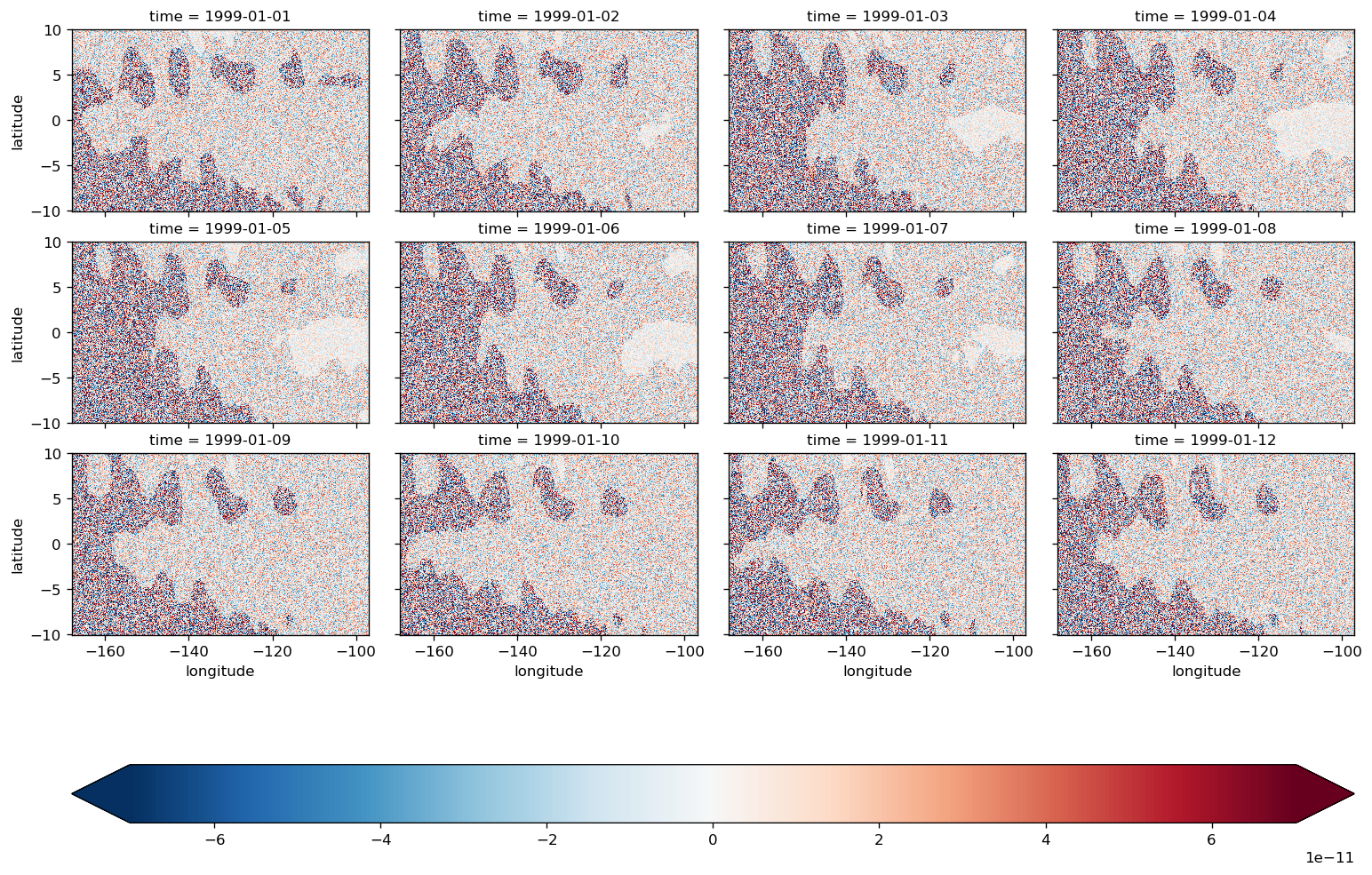
diff = (LHS - RHS).compute()
diff.mean("time").plot(robust=True, x="longitude")
plot_lhs_rhs(LHS.isel(time=10), RHS.isel(time=10))
all time steps#
plot_lhs_rhs(LHS, RHS)
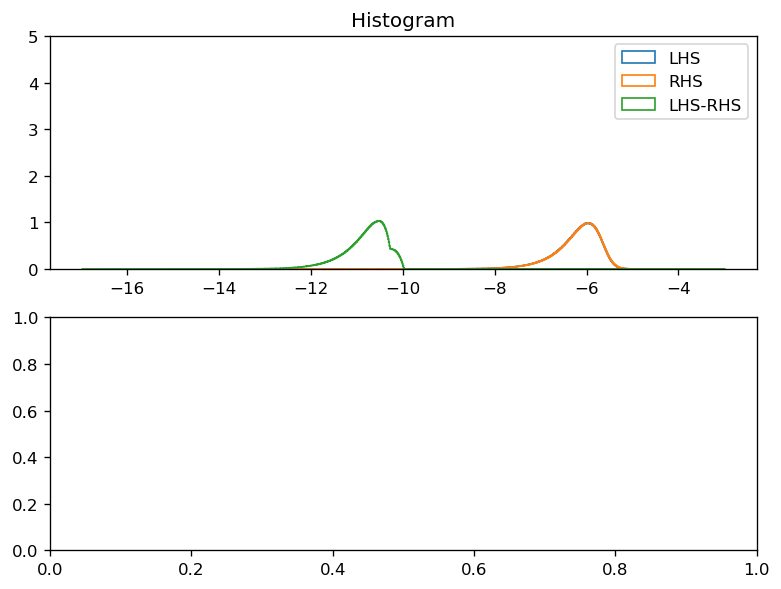
after shifting SSH one iteration back in time#
residual ~ 1e-11
diff.plot(col="time", col_wrap=4, robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x2b7671fde6a0>
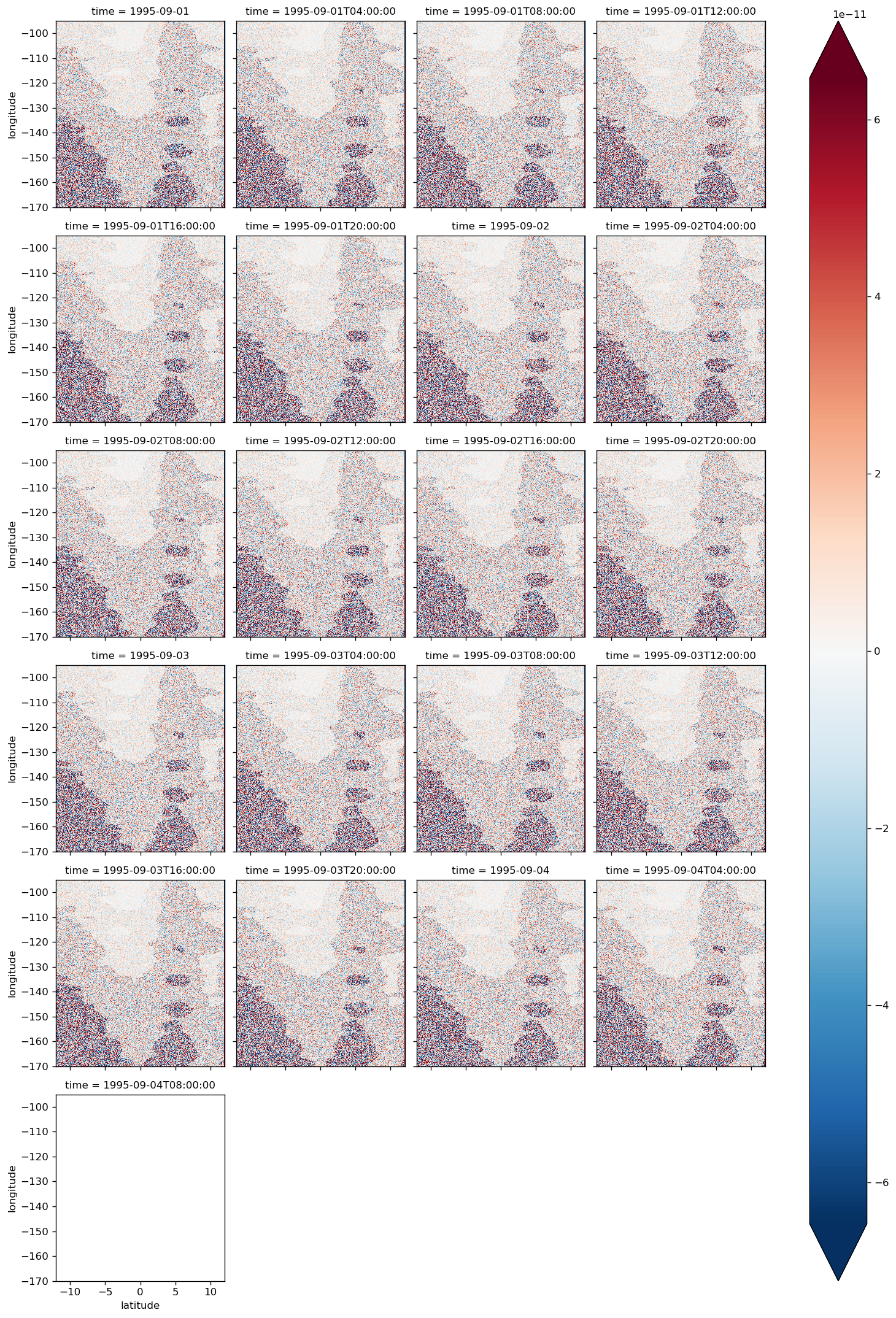
before shifting SSH one iteration back in time#
residual ~ 1e-7
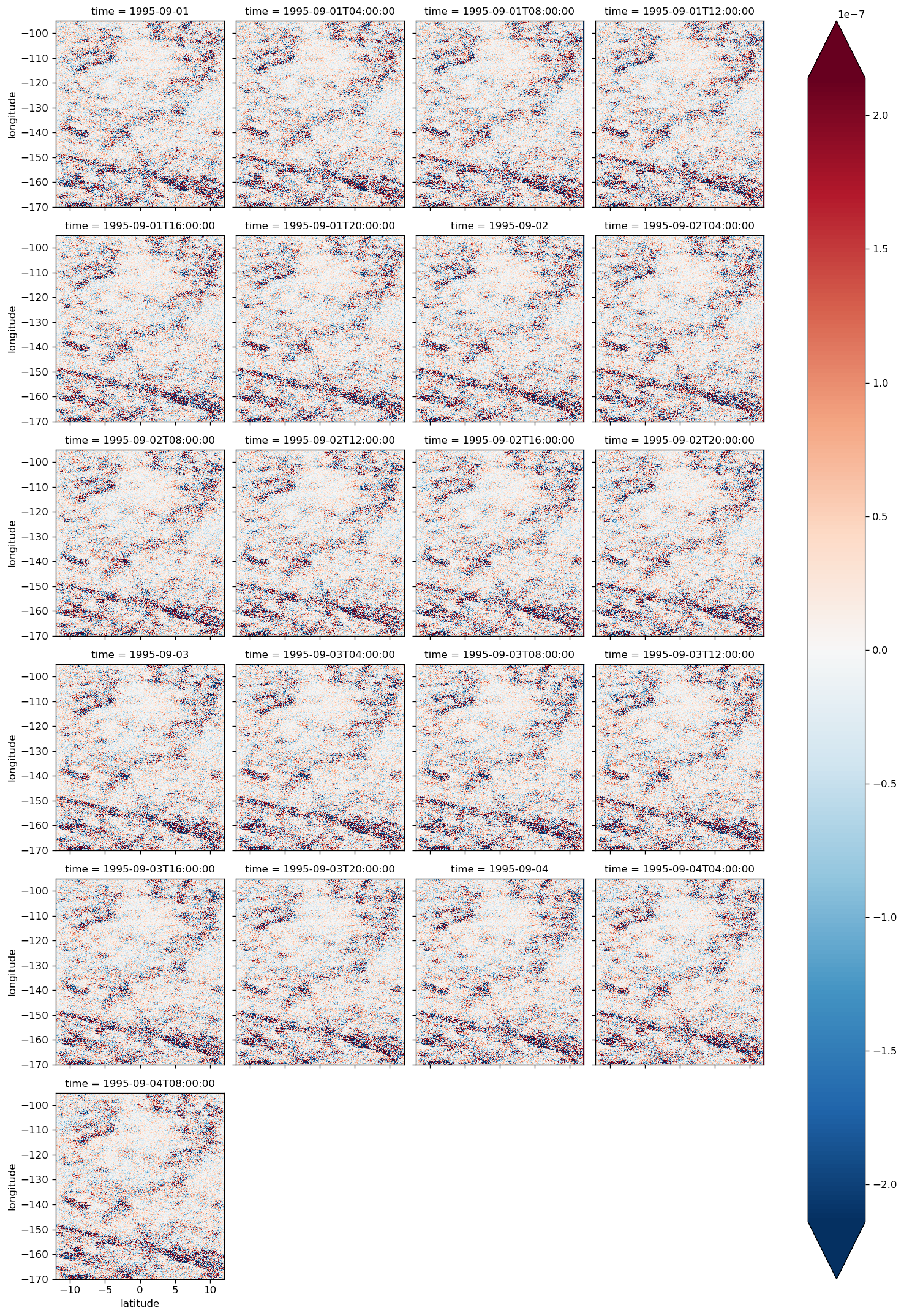
diff.plot(col="time", col_wrap=4, robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x2b7674a99978>
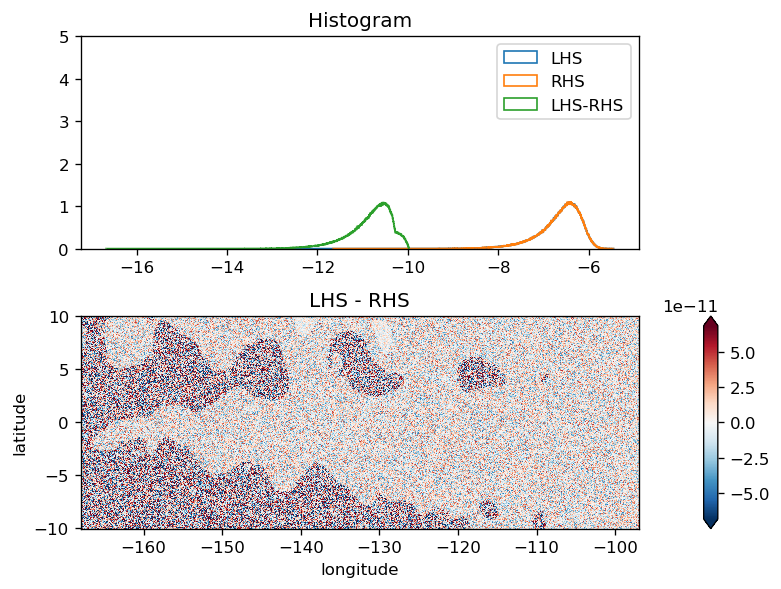
V momentum#
This seems to have some systematic error but it is small
level = 10
SSHy = (
-9.81
* budget.ETAN.diff("latitude", label="upper").reindex(latitude=budget.latitude)
/ metrics.DYC
)
# from http://mailman.mitgcm.org/pipermail/mitgcm-support/2010-December/006921.html
Vm_Impl = VISrI_Vm.diff("depth", label="lower") / (
metrics.rAw * metrics.drF.isel(depth=level) * metrics.hFacW.isel(depth=level)
)
RHS = (
SSHy
+ budget.Vm_dPHdy
+ budget.Vm_Advec
# + budget.Vm_Cori # in Vm_Advec
+ budget.Vm_Diss
+ Vm_Impl
+ budget.Vm_Ext.fillna(0)
+ budget.AB_gV
).isel(depth=level)
LHS = budget.TOTVTEND.isel(depth=level) / 86400
/glade/u/home/dcherian/miniconda3/envs/dcpy/lib/python3.6/site-packages/dask/array/core.py:3908: PerformanceWarning: Increasing number of chunks by factor of 21
**blockwise_kwargs,
(LHS - RHS).isel(time=slice(6)).compute().plot(
col="time",
col_wrap=3,
x="longitude",
cbar_kwargs={"orientation": "horizontal"},
robust=True,
)
<xarray.plot.facetgrid.FacetGrid at 0x2b8db41ff9b0>
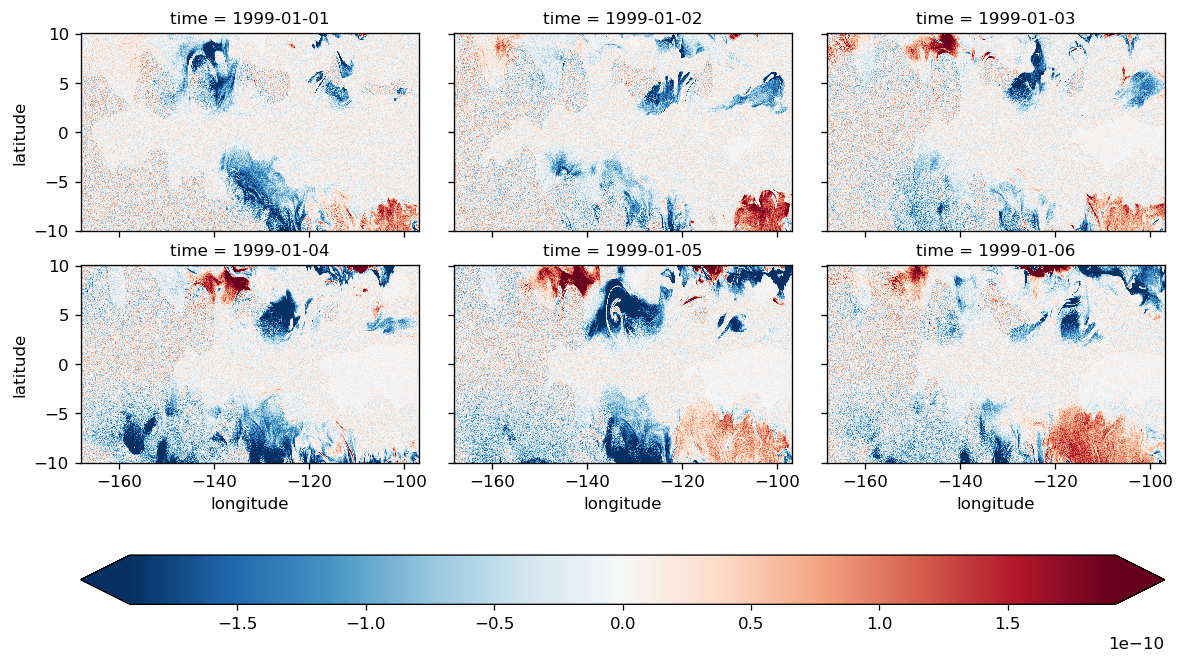
all time steps#
plot_lhs_rhs(LHS, RHS)
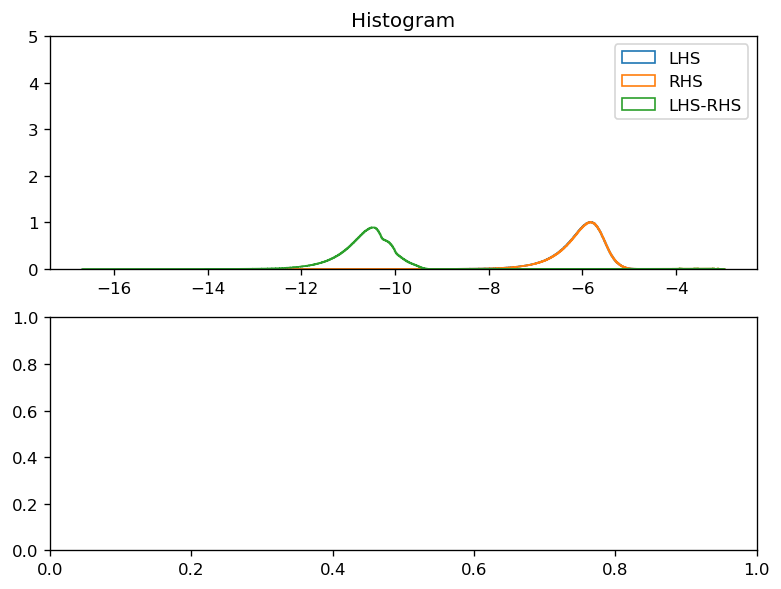
plot_lhs_rhs(LHS.isel(time=20), RHS.isel(time=20))