LES tendencies#
This has to be run on casper
I would add a 5 day buffer on
les_time_lengthI’ve removed the coriolis contribution, so the LES needs
f≠0I have not checked the 12H offset in time
/glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_runs_setup_scripts/tpos-DIABLO/diablo_2.0/tpos_config_files
import glob
import cf_xarray
import dask
import dcpy
import h5py
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import xarray as xr
import xgcm
import pump
import pump.les
import pump.model
from pump.les import interleave_time, interpolate, write_to_txt
dask.config.set(**{"array.slicing.split_large_chunks": False}) # silence some warnings
<dask.config.set at 0x2b963be83ee0>
Sets up a dask cluster; only run once
import ncar_jobqueue
cluster = ncar_jobqueue.NCARCluster(account="ncgd0011")
cluster.scale(12)
import distributed
client = distributed.Client(cluster)
client
Client
|
Cluster
|
Things you could change; if changing, rerun all cells below (Use the Run menu above)
sst = xr.open_mfdataset(
f"{gcmdir}/*buoy*.nc",
chunks={"latitude": 120, "longitude": 500, "depth": 1},
combine="by_coords",
parallel=True,
).theta.isel(depth=0)
f, axx = plt.subplots(10, 2, sharey=True, constrained_layout=True)
for year, ax in zip(np.unique(sst.time.dt.year), axx.flat):
(
sst.theta.isel(depth=0)
.sel(latitude=slice(-2, 5))
.sel(time=slice(f"{year}-Jun", f"{year}-Dec"))
.sel(longitude=-110, method="nearest")
.plot(
x="time",
ax=ax,
vmin=22,
vmax=28,
cmap=mpl.cm.RdYlBu_r,
add_colorbar=False,
add_labels=False,
)
)
f.set_size_inches((12, 18))
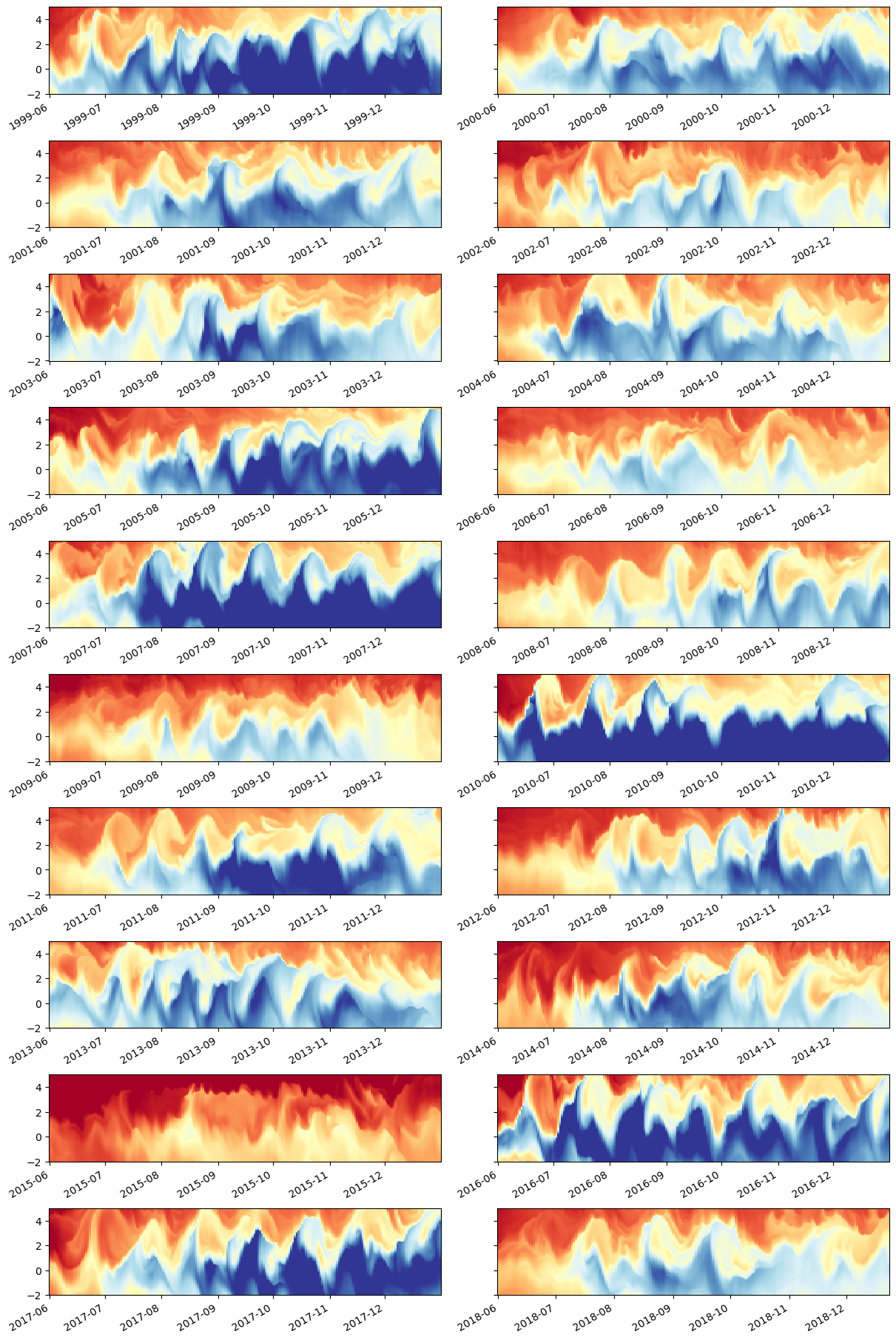
Choose time period + plot ocean state to see what we get#
gcmdir = "/glade/campaign/cgd/oce/people/bachman/TPOS_1_20_20_year_output/" # MITgcm output directory
outdir = "/glade/work/dcherian/pump/les_forcing/mitgcm_test/" # output directory for txt files
whichz = "216" # or "288" # which z grid h5 file to use
leslon = -110 # it will find the nearest
leslat = 0 # will find the nearest
# start date for les; noon is a good time (it is before sunrise)
sim_time = pd.Timestamp("2015-09-20 12:00:00")
# ADD a 5 day buffer here (:] all kinds of bugs at the beginning and end)
les_time_length = 30 # (days); length of time for forcing/pushing files
# don't change anything here
output_start_time = pd.Timestamp("1999-01-01") # don't change
firstfilenum = (
(
sim_time - pd.Timedelta("1D") - output_start_time
) # add one day offset to avoid bugs
.to_numpy()
.astype("timedelta64[D]")
.astype("int")
)
lastfilenum = firstfilenum + les_time_length
def gen_file_list(suffix):
files = [
f"{gcmdir}/File_{num}_{suffix}.nc"
for num in range(firstfilenum - 1, lastfilenum + 1)
]
return files
# station = pump.model.model.read_stations_20(
# dayglobstr=folder, dirname=gcmdir, globstr="*_-140*.nc"
# )
state = xr.open_mfdataset(
gen_file_list(suffix="buoy"),
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
).rename({"latitude": "YC", "longitude": "XC", "depth": "RC"})
state["YC"].attrs["units"] = "degrees_north"
q, r = divmod(firstfilenum, 250)
folder = f"{250*q + 1}-{250*(q+1)}"
path = f"{gcmdir}/STATION_DATA/Day_{folder}/"
station = (
xr.open_mfdataset(
sorted(glob.glob(path + f"/*_{leslon}.000.nc")),
parallel=True,
decode_times=False,
)
.sel(depth=slice(-150))
.squeeze()
)
station.time.attrs["units"] = f"seconds since {output_start_time}"
station = xr.decode_cf(station)
station["dens"] = pump.mdjwf.dens(station.salt, station.theta, np.array([0]))
station["mld"] = pump.calc.get_mld(station.dens).compute()
station
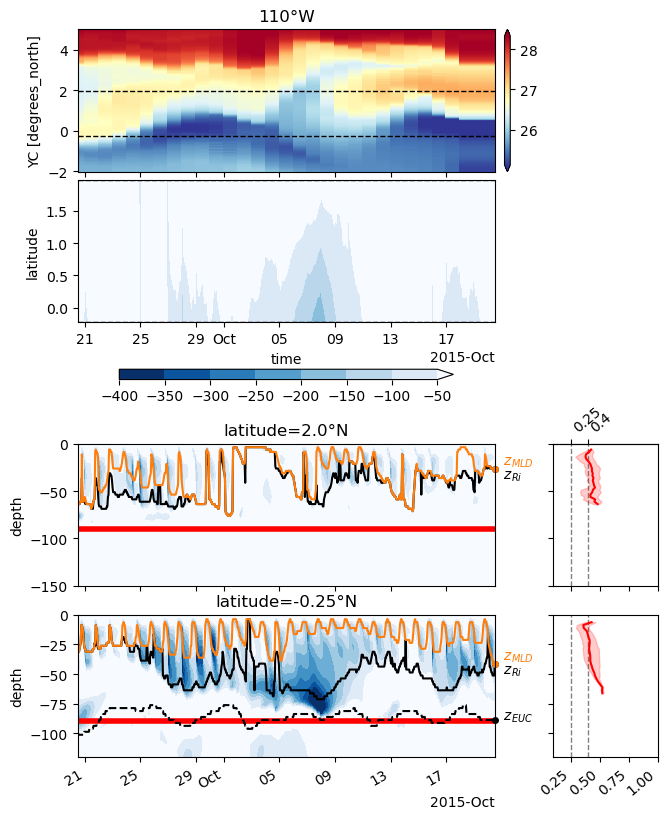
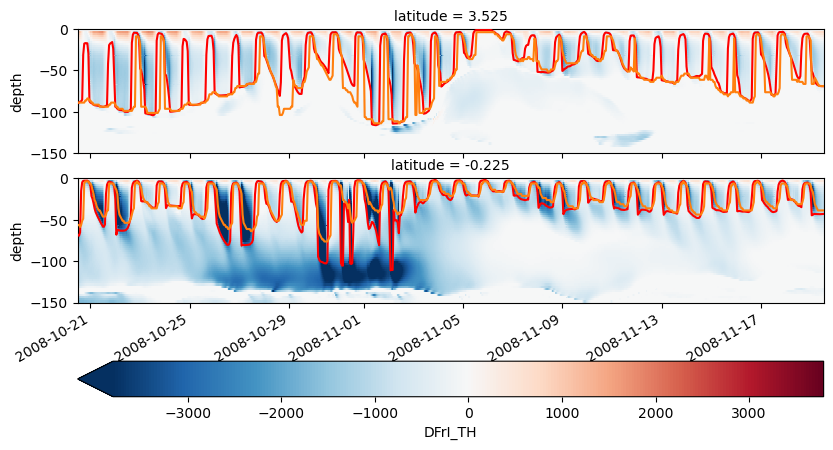
choose_lats = [-0.25, 2]
subset = station.sel(time=slice(sim_time, sim_time + pd.Timedelta("30D"))).sel(
latitude=choose_lats, method="nearest"
)
subset["sst"] = state.theta.sel(XC=leslon, method="nearest", RC=0).reindex(
time=subset.time, method="pad"
)
subset = pump.calc.calc_reduced_shear(subset)
subset["eucmax"] = pump.calc.get_euc_max(subset.isel(latitude=0).u)
subset["dcl_base"] = pump.calc.get_dcl_base_Ri(subset)
subset["Jq"] = 1035 * 3995 * subset.DFrI_TH / 3.09e7
f, ax = pump.plot.plot_jq_sst(
subset,
full=subset,
lon=leslon,
lat=choose_lats,
periods=slice(subset.time[0].values, subset.time[-1].values),
)
ax[2, 0].set_ylim([-150, 0])
calc uz
calc vz
calc S2
calc N2
calc shred2
Calc Ri
(-150.0, 0.0)
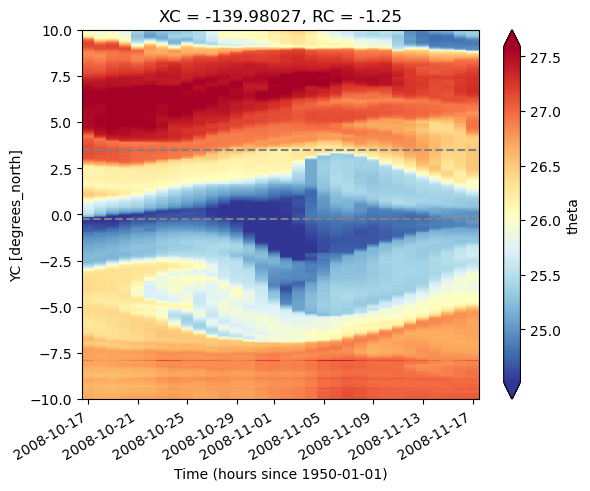
state.theta.sel(XC=leslon, method="nearest", RC=0).plot(
x="time", robust=True, cmap=mpl.cm.RdYlBu_r
)
dcpy.plots.liney(choose_lats, zorder=1)
fg = subset.DFrI_TH.sortby("latitude", ascending=False).plot(
row="latitude", robust=True, cbar_kwargs={"orientation": "horizontal"}
)
[
(-1 * subset.KPPhbl).sel(loc).plot(color="r", ax=ax, zorder=12, _labels=False)
for loc, ax in zip(fg.name_dicts.squeeze(), fg.axes.squeeze())
]
[
(subset.mld).sel(loc).plot(color="C1", ax=ax, zorder=13, _labels=False)
for loc, ax in zip(fg.name_dicts.squeeze(), fg.axes.squeeze())
]
fg.fig.set_size_inches((10, 5))
LES config#
LES things that you need not change; Grid is chosen by whichz in the above cell
# Should not need to change anything below this
griddir = "/glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_runs_setup_scripts/tpos-DIABLO/diablo_2.0/pre_process/"
renamer = {
"u": "U",
"v": "W",
"temp": "T",
# "salt": "S"
}
offset = xr.Dataset({"U": 0.4, "T": 25, "S": 35.25, "W": 0})
t0 = (sim_time).to_numpy()
grid = h5py.File(f"{griddir}/gridtpos{whichz}.h5", mode="r")
zles = grid["grids"]["y"][:]
grid.close()
zles = 0.5 * (zles[1:-1] + zles[2:])
newz = xr.DataArray(
zles - zles.max() - 0.25, dims="z", name="z" # this is what dan does
)
newz # prints out the grid
<xarray.DataArray 'z' (z: 216)>
array([-107.75, -107.25, -106.75, -106.25, -105.75, -105.25, -104.75,
-104.25, -103.75, -103.25, -102.75, -102.25, -101.75, -101.25,
-100.75, -100.25, -99.75, -99.25, -98.75, -98.25, -97.75,
-97.25, -96.75, -96.25, -95.75, -95.25, -94.75, -94.25,
-93.75, -93.25, -92.75, -92.25, -91.75, -91.25, -90.75,
-90.25, -89.75, -89.25, -88.75, -88.25, -87.75, -87.25,
-86.75, -86.25, -85.75, -85.25, -84.75, -84.25, -83.75,
-83.25, -82.75, -82.25, -81.75, -81.25, -80.75, -80.25,
-79.75, -79.25, -78.75, -78.25, -77.75, -77.25, -76.75,
-76.25, -75.75, -75.25, -74.75, -74.25, -73.75, -73.25,
-72.75, -72.25, -71.75, -71.25, -70.75, -70.25, -69.75,
-69.25, -68.75, -68.25, -67.75, -67.25, -66.75, -66.25,
-65.75, -65.25, -64.75, -64.25, -63.75, -63.25, -62.75,
-62.25, -61.75, -61.25, -60.75, -60.25, -59.75, -59.25,
-58.75, -58.25, -57.75, -57.25, -56.75, -56.25, -55.75,
-55.25, -54.75, -54.25, -53.75, -53.25, -52.75, -52.25,
-51.75, -51.25, -50.75, -50.25, -49.75, -49.25, -48.75,
-48.25, -47.75, -47.25, -46.75, -46.25, -45.75, -45.25,
-44.75, -44.25, -43.75, -43.25, -42.75, -42.25, -41.75,
-41.25, -40.75, -40.25, -39.75, -39.25, -38.75, -38.25,
-37.75, -37.25, -36.75, -36.25, -35.75, -35.25, -34.75,
-34.25, -33.75, -33.25, -32.75, -32.25, -31.75, -31.25,
-30.75, -30.25, -29.75, -29.25, -28.75, -28.25, -27.75,
-27.25, -26.75, -26.25, -25.75, -25.25, -24.75, -24.25,
-23.75, -23.25, -22.75, -22.25, -21.75, -21.25, -20.75,
-20.25, -19.75, -19.25, -18.75, -18.25, -17.75, -17.25,
-16.75, -16.25, -15.75, -15.25, -14.75, -14.25, -13.75,
-13.25, -12.75, -12.25, -11.75, -11.25, -10.75, -10.25,
-9.75, -9.25, -8.75, -8.25, -7.75, -7.25, -6.75,
-6.25, -5.75, -5.25, -4.75, -4.25, -3.75, -3.25,
-2.75, -2.25, -1.75, -1.25, -0.75, -0.25])
Dimensions without coordinates: z- z: 216
- -107.8 -107.2 -106.8 -106.2 -105.8 ... -2.25 -1.75 -1.25 -0.75 -0.25
array([-107.75, -107.25, -106.75, -106.25, -105.75, -105.25, -104.75, -104.25, -103.75, -103.25, -102.75, -102.25, -101.75, -101.25, -100.75, -100.25, -99.75, -99.25, -98.75, -98.25, -97.75, -97.25, -96.75, -96.25, -95.75, -95.25, -94.75, -94.25, -93.75, -93.25, -92.75, -92.25, -91.75, -91.25, -90.75, -90.25, -89.75, -89.25, -88.75, -88.25, -87.75, -87.25, -86.75, -86.25, -85.75, -85.25, -84.75, -84.25, -83.75, -83.25, -82.75, -82.25, -81.75, -81.25, -80.75, -80.25, -79.75, -79.25, -78.75, -78.25, -77.75, -77.25, -76.75, -76.25, -75.75, -75.25, -74.75, -74.25, -73.75, -73.25, -72.75, -72.25, -71.75, -71.25, -70.75, -70.25, -69.75, -69.25, -68.75, -68.25, -67.75, -67.25, -66.75, -66.25, -65.75, -65.25, -64.75, -64.25, -63.75, -63.25, -62.75, -62.25, -61.75, -61.25, -60.75, -60.25, -59.75, -59.25, -58.75, -58.25, -57.75, -57.25, -56.75, -56.25, -55.75, -55.25, -54.75, -54.25, -53.75, -53.25, -52.75, -52.25, -51.75, -51.25, -50.75, -50.25, -49.75, -49.25, -48.75, -48.25, -47.75, -47.25, -46.75, -46.25, -45.75, -45.25, -44.75, -44.25, -43.75, -43.25, -42.75, -42.25, -41.75, -41.25, -40.75, -40.25, -39.75, -39.25, -38.75, -38.25, -37.75, -37.25, -36.75, -36.25, -35.75, -35.25, -34.75, -34.25, -33.75, -33.25, -32.75, -32.25, -31.75, -31.25, -30.75, -30.25, -29.75, -29.25, -28.75, -28.25, -27.75, -27.25, -26.75, -26.25, -25.75, -25.25, -24.75, -24.25, -23.75, -23.25, -22.75, -22.25, -21.75, -21.25, -20.75, -20.25, -19.75, -19.25, -18.75, -18.25, -17.75, -17.25, -16.75, -16.25, -15.75, -15.25, -14.75, -14.25, -13.75, -13.25, -12.75, -12.25, -11.75, -11.25, -10.75, -10.25, -9.75, -9.25, -8.75, -8.25, -7.75, -7.25, -6.75, -6.25, -5.75, -5.25, -4.75, -4.25, -3.75, -3.25, -2.75, -2.25, -1.75, -1.25, -0.75, -0.25])
MITgcm#
The daily avg output was subset in MATLAB syntax : 41:end-40, 41:end-40, 1:nz
Read all ocean model output#
coords = pump.model.read_mitgcm_coords(gcmdir)
grid = xgcm.Grid(
coords, periodic=False, boundary={"X": "extend", "Y": "extend", "Z": "extend"}
)
# ssh data
ssh = (
xr.open_mfdataset(
gen_file_list(suffix="etan"),
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
)
.rename({"latitude": "YC", "longitude": "XC"})
.update(coords.coords)
)
# momentum budget terms
budgets = xr.open_mfdataset(
gen_file_list(suffix="ub") + gen_file_list(suffix="vb"),
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
)
budgets = pump.model.rename_mitgcm_budget_terms(budgets, coords).update(coords.coords)
# surface data
surf = (
xr.open_mfdataset(
gen_file_list(suffix="surf"),
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
)
.rename({"latitude": "YC", "longitude": "XC"})
.update(coords.coords)
)
surf["oceQsw"] = surf.oceQsw.fillna(0)
surf["taux"] = 1035 * 2.5 * budgets["Um_Ext"].isel(RC=0)
surf["tauy"] = 1035 * 2.5 * budgets["Vm_Ext"].isel(RC=0)
# heat budget terms
hb = xr.open_mfdataset(
gen_file_list(suffix="hb"),
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
)
hb_ren = pump.model.rename_mitgcm_budget_terms(hb, coords).update(coords.coords)
# state variables
state = xr.open_mfdataset(
gen_file_list(suffix="buoy"),
chunks={"latitude": 120, "longitude": 500},
combine="by_coords",
parallel=True,
).rename({"latitude": "YC", "longitude": "XC", "depth": "RC"})
state["u"] = state.u.rename({"XC": "XG"})
state["v"] = state.v.rename({"YC": "YG"})
state = state.update(coords.coords)
# grid metrics
metrics = (
pump.model.read_metrics(gcmdir)
.compute()
.isel(longitude=slice(40, -40), latitude=slice(40, -40), depth=slice(136))
.chunk({"latitude": 120, "longitude": 500})
.drop(["latitude", "longitude", "depth"])
.rename({"depth_left": "RF", "depth": "RC", "latitude": "YC", "longitude": "XC"})
.set_coords("RF")
.isel(RF=slice(136))
)
metrics["rAw"] = metrics.rAw.rename({"XC": "XG"})
metrics["rAs"] = metrics.rAs.rename({"YC": "YG"})
metrics["hFacW"] = metrics.hFacW.rename({"XC": "XG"})
metrics["hFacS"] = metrics.hFacS.rename({"YC": "YG"})
metrics["DXG"] = metrics.DXG.rename({"YC": "YG"})
metrics["DYG"] = metrics.DYG.rename({"XC": "XG"})
metrics = metrics.update(coords.coords)
# penetrative shortwave radiation profile
swprofile = (
xr.DataArray(
pump.model.mitgcm_sw_prof(metrics.RF[:-1].values)
- pump.model.mitgcm_sw_prof(metrics.RF[1:].values),
dims=["RC"],
attrs={"long_name": "SW radiation deposited in cell"},
)
.assign_coords(RC=coords.coords["RC"][:-1])
.reindex(RC=coords.coords["RC"])
)
Offset in time (I have not checked this).
ssh["time"] = ssh.time + pd.Timedelta("12H")
budgets["time"] = budgets.time + pd.Timedelta("12H")
state["time"] = state.time + pd.Timedelta("12h")
hb_ren["time"] = hb_ren.time + pd.Timedelta("12h")
surf["time"] = surf.time + pd.Timedelta("12h")
Make sure that everything is on the same grid; just a check.
xr.align(budgets, ssh, state, hb_ren, metrics, surf, join="exact");
Estimate flux divergences#
# constants taken from diagnostics.log (I think)
global_area = 2.196468634481708e13
rhoConst = 1035
Cp = 3994
f0 = 2 * (2 * np.pi / 86400) * np.sin(leslat * np.pi / 180)
# This is WRONG! Because we have subset the output this won't recover the MITgcm correction exactly
# seems to be particularly bad at t=0
surf_mass = (hb_ren.WTHMASS * metrics.RAC).isel(RC=0)
TsurfCor = surf_mass.sum(["XC", "YC"]) / global_area
def expand_depth(ds):
if "RC" not in ds:
ds["RC"] = coords.RC.isel(RC=0).data
return ds.expand_dims("RC").reindex(RC=coords.RC, fill_value=0)
budgets["surf_corr_tend"] = -1 * (
(TsurfCor - hb_ren.WTHMASS.isel(RC=0))
/ metrics.dRF.isel(RC=0)
/ metrics.hFacC.isel(RC=0)
).pipe(expand_depth)
budgets["TOTTTEND"] = hb_ren.TOTTTEND
budgets["ADV_TH"] = (
-1
* (
grid.diff(hb_ren.ADVx_TH, "X")
+ grid.diff(hb_ren.ADVy_TH, "Y")
- grid.diff(
hb_ren.ADVr_TH.fillna(0), "Z"
) # negative because level 0 is surface
)
/ metrics.cellvol
)
budgets["MIX_TH"] = (
grid.diff(hb_ren.DFrI_TH.fillna(0) + hb_ren.KPPg_TH.fillna(0), "Z")
/ metrics.cellvol
)
shfluxminusswrad = (
-1
* (surf.TFLUX - surf.oceQsw)
/ (rhoConst * Cp * metrics.dRF.isel(RC=0) * metrics.hFacC.isel(RC=0))
)
SW = (
-1 * surf.oceQsw / (rhoConst * Cp) / (metrics.dRF[0] * metrics.hFacC[0]) * swprofile
)
# residual is LHS - RHS
# this is non-zero because of some global correction at the surface
budgets["RESIDUAL_TH"] = -1 * expand_depth(
(
shfluxminusswrad
+ (budgets.surf_corr_tend + budgets.MIX_TH + budgets.ADV_TH + SW).isel(RC=0)
)
- budgets.TOTTTEND.isel(RC=0) / 86400
)
budgets["SSHx"] = -9.81 * grid.diff(ssh.ETAN, "X") / metrics.DXC.data
budgets["SSHy"] = -9.81 * grid.diff(ssh.ETAN, "Y") / metrics.DYC.data
budgets["Um_Impl"] = (
grid.diff(budgets.VISrI_Um.fillna(0), "Z")
/ metrics.rAw
/ metrics.drF
/ metrics.hFacW
)
budgets["Vm_Impl"] = (
grid.diff(budgets.VISrI_Vm.fillna(0), "Z")
/ metrics.rAs
/ metrics.drF
/ metrics.hFacS
)
# Has LHS sign because we want to subtract it from the Advective part
budgets["Um_cor"] = -1 * (+1 * f0 * grid.interp(state.v, ("X", "Y")))
budgets["Vm_cor"] = -1 * (-1 * f0 * grid.interp(state.u, ("X", "Y")))
budgets = budgets.cf.guess_coord_axis()
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.8/site-packages/dask/array/core.py:377: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
o = func(*args, **kwargs)
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.8/site-packages/dask/array/core.py:377: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
o = func(*args, **kwargs)
Tendency terms#
Now subset the RHS terms to domain; add sum to total RHS tendencies.
Stupidly, the temperature flux divergences in budgets are with LHS sign; so invert that at the end
subrhs = subsetter(budgets)
rhs = xr.Dataset()
# these are names of variables in budgets that are added together in the for loop
var_terms = {
"u": ["Um_dPHdx", "Um_Advec", "Um_Diss", "AB_gU", "SSHx", "Um_cor"],
"v": ["Vm_dPHdy", "Vm_Advec", "Vm_Diss", "AB_gV", "SSHy", "Vm_cor"],
"temp": ["ADV_TH", "surf_corr_tend", "RESIDUAL_TH"],
}
for var, varnames in var_terms.items():
rhs[var] = (
subrhs[varnames].pipe(interpolate, newz).to_array("term").sum("term")
) * 86400
rhs[var].attrs["description"] = f"sum over {varnames}"
rhs
<xarray.Dataset>
Dimensions: (RC: 216, time: 28)
Coordinates:
XG float64 -140.0
XC float64 -140.0
YG float64 3.607e-14
YC float64 0.025
* time (time) datetime64[ns] 2008-10-24T12:00:00 ... 2008-11-20T12:00:00
* RC (RC) float64 -107.8 -107.2 -106.8 -106.2 ... -1.25 -0.75 -0.25
Data variables:
u (time, RC) float64 -0.0001335 -0.0001465 ... 0.07101 0.07279
v (time, RC) float64 -0.0348 -0.03506 -0.03533 ... 0.007359 0.006686
temp (time, RC) float64 0.1456 0.1421 0.1386 ... -0.04826 -0.05021- RC: 216
- time: 28
- XG()float64-140.0
- axis :
- X
- c_grid_axis_shift :
- -0.5
array(-139.97500076)
- XC()float64-140.0
- axis :
- X
array(-140.00000038)
- YG()float643.607e-14
- axis :
- Y
- c_grid_axis_shift :
- -0.5
array(3.60683705e-14)
- YC()float640.025
- axis :
- Y
array(0.025)
- time(time)datetime64[ns]2008-10-24T12:00:00 ... 2008-11-...
- axis :
- T
- standard_name :
- time
array(['2008-10-24T12:00:00.000000000', '2008-10-25T12:00:00.000000000', '2008-10-26T12:00:00.000000000', '2008-10-27T12:00:00.000000000', '2008-10-28T12:00:00.000000000', '2008-10-29T12:00:00.000000000', '2008-10-30T12:00:00.000000000', '2008-10-31T12:00:00.000000000', '2008-11-01T12:00:00.000000000', '2008-11-02T12:00:00.000000000', '2008-11-03T12:00:00.000000000', '2008-11-04T12:00:00.000000000', '2008-11-05T12:00:00.000000000', '2008-11-06T12:00:00.000000000', '2008-11-07T12:00:00.000000000', '2008-11-08T12:00:00.000000000', '2008-11-09T12:00:00.000000000', '2008-11-10T12:00:00.000000000', '2008-11-11T12:00:00.000000000', '2008-11-12T12:00:00.000000000', '2008-11-13T12:00:00.000000000', '2008-11-14T12:00:00.000000000', '2008-11-15T12:00:00.000000000', '2008-11-16T12:00:00.000000000', '2008-11-17T12:00:00.000000000', '2008-11-18T12:00:00.000000000', '2008-11-19T12:00:00.000000000', '2008-11-20T12:00:00.000000000'], dtype='datetime64[ns]') - RC(RC)float64-107.8 -107.2 ... -0.75 -0.25
array([-107.75, -107.25, -106.75, ..., -1.25, -0.75, -0.25])
- u(time, RC)float64-0.0001335 -0.0001465 ... 0.07279
- description :
- sum over ['Um_dPHdx', 'Um_Advec', 'Um_Diss', 'AB_gU', 'SSHx', 'Um_cor']
array([[-1.33460647e-04, -1.46488235e-04, -1.59515824e-04, ..., 5.24772145e-02, 5.28543083e-02, 5.32314022e-02], [ 9.57218729e-03, 9.52312693e-03, 9.47406658e-03, ..., -1.96129674e-02, -1.96432441e-02, -1.96735208e-02], [-3.79550307e-02, -3.84084858e-02, -3.88619410e-02, ..., -7.41253959e-02, -7.41008169e-02, -7.40762379e-02], ..., [ 8.72896919e-02, 8.54605338e-02, 8.36313757e-02, ..., 1.30402331e-01, 1.30762128e-01, 1.31121925e-01], [-6.28008741e-02, -6.38680041e-02, -6.49351340e-02, ..., 9.87972187e-02, 9.99107269e-02, 1.01024235e-01], [ 1.57595680e-01, 1.50550090e-01, 1.43504499e-01, ..., 6.92405788e-02, 7.10133945e-02, 7.27862102e-02]]) - v(time, RC)float64-0.0348 -0.03506 ... 0.006686
- description :
- sum over ['Vm_dPHdy', 'Vm_Advec', 'Vm_Diss', 'AB_gV', 'SSHy', 'Vm_cor']
array([[-0.03480268, -0.03506462, -0.03532655, ..., -0.12910898, -0.13012529, -0.13114159], [-0.05375721, -0.05411227, -0.05446733, ..., -0.15080738, -0.15209924, -0.15339111], [-0.04946468, -0.04981234, -0.05016 , ..., -0.1262247 , -0.1275586 , -0.12889251], ..., [ 0.05728015, 0.05904402, 0.0608079 , ..., -0.04430217, -0.043594 , -0.04288583], [ 0.09898644, 0.0992291 , 0.09947175, ..., -0.02428349, -0.02447475, -0.02466601], [ 0.01163478, 0.0183144 , 0.02499402, ..., 0.00803147, 0.00735881, 0.00668614]]) - temp(time, RC)float640.1456 0.1421 ... -0.04826 -0.05021
- description :
- sum over ['ADV_TH', 'surf_corr_tend', 'RESIDUAL_TH']
array([[ 0.14564553, 0.14213041, 0.13861529, ..., -0.02581352, -0.02724174, -0.02866996], [-0.00715235, -0.0053795 , -0.00360665, ..., -0.05635626, -0.05779917, -0.05924209], [ 0.1345301 , 0.13257305, 0.130616 , ..., -0.06998087, -0.07188222, -0.07378356], ..., [-0.6414084 , -0.59282341, -0.54423842, ..., -0.01810363, -0.01898098, -0.01985832], [ 0.61336345, 0.59662818, 0.57989292, ..., 0.04802301, 0.0463206 , 0.0446182 ], [-1.64504769, -1.50867446, -1.37230123, ..., -0.04632038, -0.04826419, -0.05020799]])
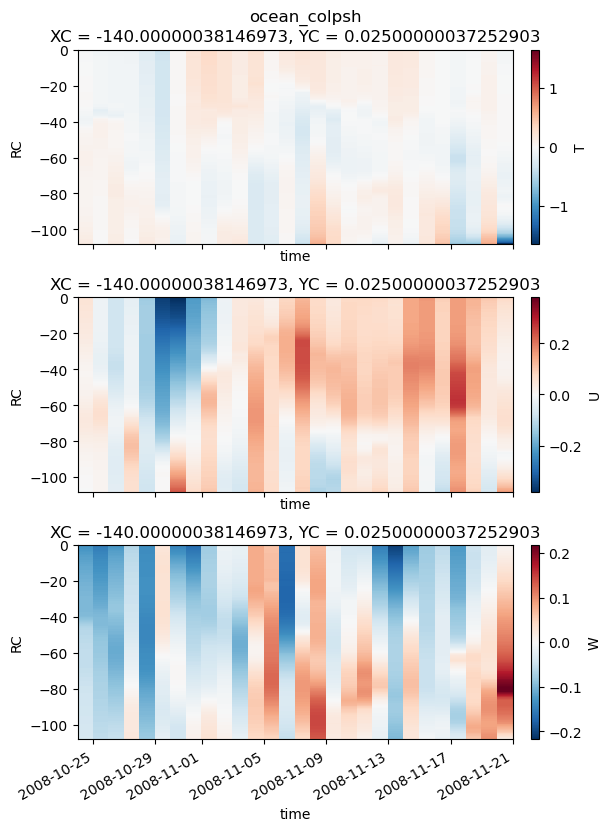
write_to_txt(
rhs.rename(renamer).compute().sel(time=slice(t0, None)),
outdir=outdir,
prefix="ocean_colpsh_",
interleave=True,
t0=t0,
)
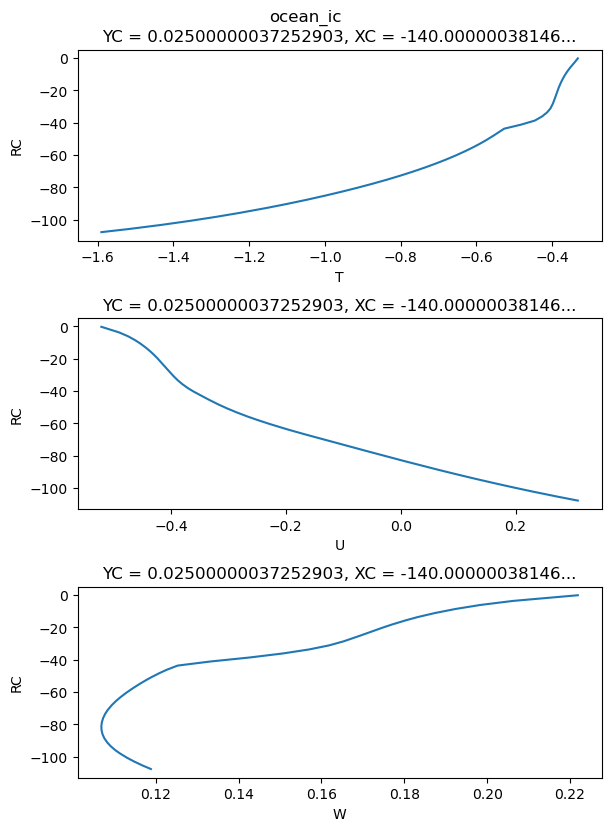
initial condition#
subds = (
state[["u", "v", "theta"]]
.rename({"theta": "temp"})
.pipe(subsetter)
.pipe(interpolate, newz)
.rename(renamer)
)
substate = (subds[["U", "W", "T"]].compute()) - offset
ic = substate.cf.interp(time=pd.Timestamp(t0))
write_to_txt(ic.compute(), outdir=outdir, prefix="ocean_ic_", interleave=False)
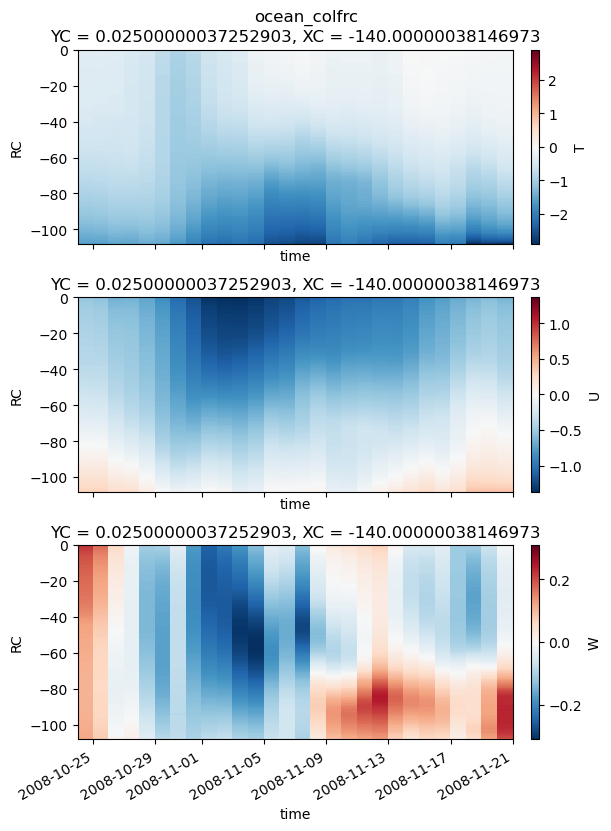
restoring#
write_to_txt(substate, outdir=outdir, prefix="ocean_colfrc_", interleave=True, t0=t0)
Estimate surface forcing from JRA using coare3.6#
jrafull = pump.obs.read_jra_20()
# undo the local time correction for LES stuff
jrafull["time"] = jrafull.time + pd.Timedelta("7h")
# interpolate to location + subset in time
jrai = (
jrafull.sel(
time=slice(
t0 - pd.Timedelta("2D"), # something is buggy so start an extra day early
substate.time[-1] + pd.Timedelta("23H"),
)
)
.interp(latitude=substate.YC.data, longitude=substate.XC.data)
.compute()
.interpolate_na("time")
)
if jrai.sizes["time"] == 0:
raise ValueError("JRA Dataset does not overlap with simulation time period!")
# calculate coare3.6 fluxes
coare, _ = pump.coare_fluxes_jra(
subsetter(state, time=False).drop(["XG", "YG"]).interp(time=jrai.time.data),
forcing=jrai,
)
coare.load()
daily_coare = coare.resample(time="D", loffset="12H").mean()
Estimate daily residuals
flux_residuals = xr.Dataset()
flux_residuals["short"] = daily_coare.short - surf["oceQsw"].pipe(subsetter, time=False)
flux_residuals["net"] = daily_coare.netflux - surf["TFLUX"].pipe(subsetter, time=False)
flux_residuals["taux"] = daily_coare.taux - surf.taux.pipe(subsetter, time=False)
flux_residuals["tauy"] = daily_coare.tauy - surf.tauy.pipe(subsetter, time=False)
flux_residuals = flux_residuals.reindex(
time=coare.time.sel(
time=slice(t0 - pd.Timedelta("1D"), t0 + pd.Timedelta(f"{les_time_length-3}D"))
),
method="nearest",
).load()
for k, v in flux_residuals.isnull().sum().compute().items():
assert v == 0, "NaNs in flux_residuals"
f = pump.les.debug_plots(flux_residuals)
f.savefig(f"{outdir}/coare_flux_residuals.png", dpi=150)
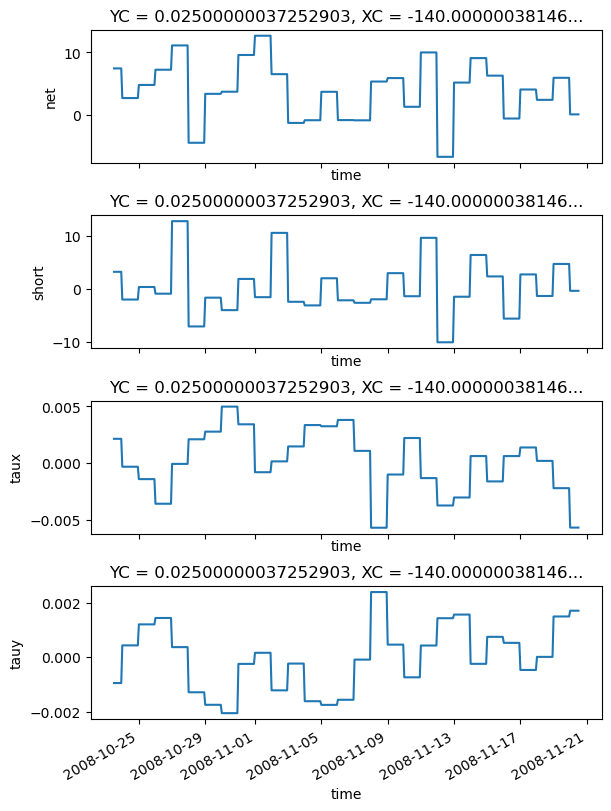
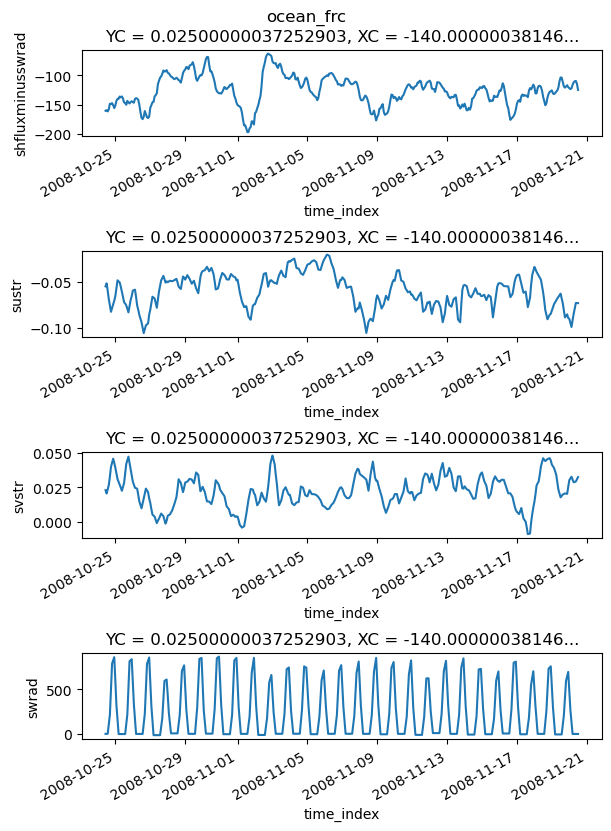
frc = xr.Dataset()
frc["sustr"] = coare.taux - flux_residuals.taux # ρ * Δz * external forcing term
frc["svstr"] = coare.tauy - flux_residuals.tauy
frc["swrad"] = coare.short - flux_residuals.short
frc["shfluxminusswrad"] = (coare.netflux - coare.short) - (
flux_residuals.net - flux_residuals.short
)
frc = frc.sel(time=slice(t0 - pd.Timedelta("1.5H"), None))
frc = frc.rename({"time": "time_index"})
time = frc.cf.indexes["time_index"].to_numpy()
frc["time"] = ("time_index", (time - t0).astype("timedelta64[s]").astype(float))
frc["time_index"].attrs["axis"] = "T"
frc.load()
write_to_txt(frc, outdir=outdir, prefix="ocean_frc_", interleave=False)
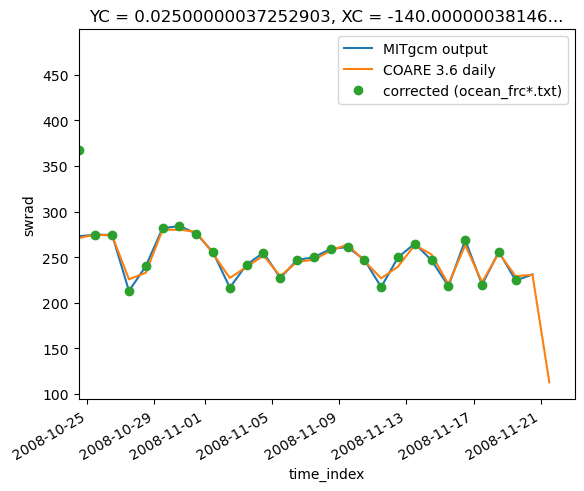
Test: coare fluxes vs mitgcm#
(surf["oceQsw"].pipe(subsetter)).plot()
daily_coare.short.plot()
(frc["swrad"].cf.resample(T="D", loffset="12H").mean()[:-1]).plot(marker="o", ls="none")
plt.legend(["MITgcm output", "COARE 3.6 daily", "corrected (ocean_frc*.txt)"])
plt.xlim((t0, None))
(14176.5, 14206.0)
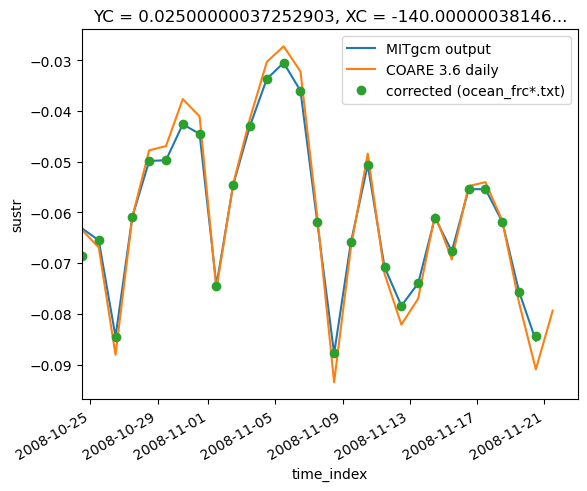
surf.taux.pipe(subsetter).plot()
daily_coare.taux.plot()
frc.cf.resample(T="D", loffset="12H").mean().sustr.plot(marker="o", ls="none")
plt.legend(["MITgcm output", "COARE 3.6 daily", "corrected (ocean_frc*.txt)"])
plt.xlim((t0, None))
(14176.5, 14206.0)
surf.tauy.pipe(subsetter).plot()
daily_coare.tauy.plot()
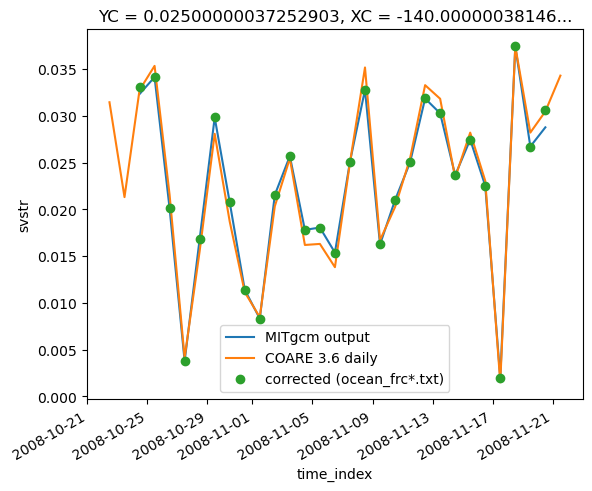
frc.cf.resample(T="D", loffset="12H").mean().svstr.plot(marker="o", ls="none")
plt.legend(["MITgcm output", "COARE 3.6 daily", "corrected (ocean_frc*.txt)"])
<matplotlib.legend.Legend at 0x2b56747a20a0>
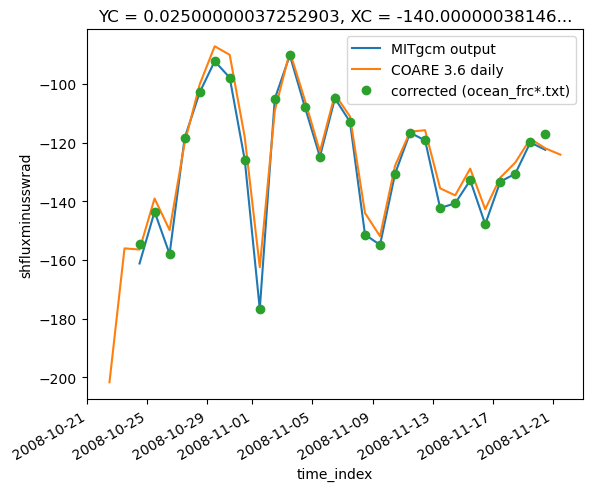
(surf.TFLUX - surf.oceQsw).pipe(subsetter).plot()
(daily_coare.netflux - daily_coare.short).plot()
frc.shfluxminusswrad.cf.resample(T="D", loffset="12H").mean().plot(
marker="o", ls="none"
)
plt.legend(["MITgcm output", "COARE 3.6 daily", "corrected (ocean_frc*.txt)"])
<matplotlib.legend.Legend at 0x2b5674a304f0>
Raise error here so you don’t go past if running every cell :)
stop here
File "<ipython-input-14-a96ba3aab008>", line 1
stop here
^
SyntaxError: invalid syntax
Debugging checks#
Test: heat budget#
LHS = budgets.TOTTTEND / 86400
RHS = (
expand_depth(shfluxminusswrad)
+ budgets.surf_corr_tend
+ budgets.MIX_TH
+ budgets.ADV_TH
+ SW
+ budgets.RESIDUAL_TH
)
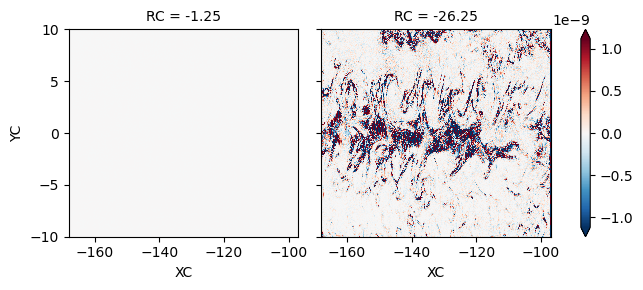
# t =0 heat budget is weird at the surface
(LHS - RHS).isel(time=1, RC=[0, 10]).plot(robust=True, col="RC")
/glade/u/home/dcherian/miniconda3/envs/dcpy/lib/python3.8/site-packages/dask/array/core.py:377: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
o = func(*args, **kwargs)
<xarray.plot.facetgrid.FacetGrid at 0x2abcfc9f3610>
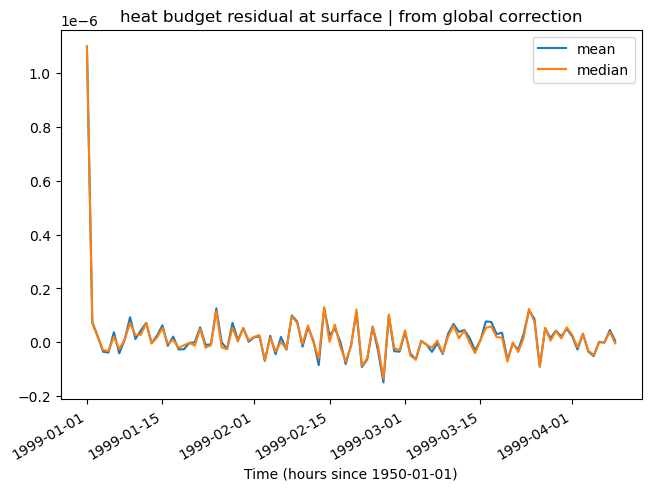
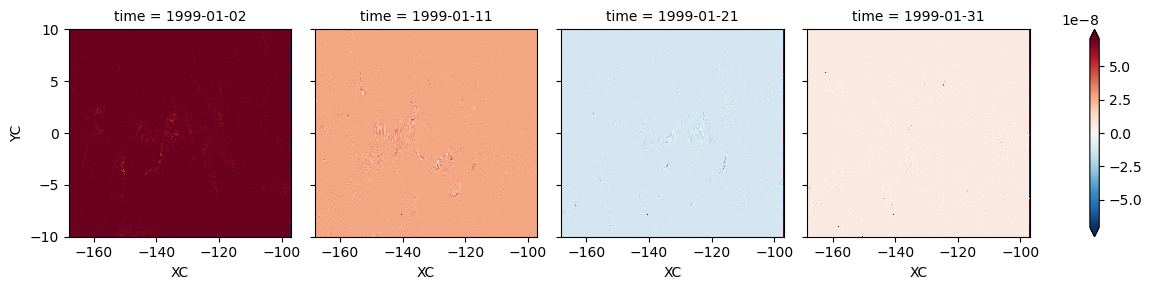
residual = (LHS - RHS).isel(RC=0)
resmedian = residual.chunk({"time": 1}).median(["XC", "YC"]).compute()
rest = residual.mean(["XC", "YC"]).compute()
import matplotlib.pyplot as plt
plt.figure(constrained_layout=True)
rest.plot()
resmedian.plot()
plt.legend(["mean", "median"])
plt.title("heat budget residual at surface | from global correction")
plt.savefig("../images/heat-budget-residual.png")
residual.isel(time=[1, 10, 20, 30]).plot(col="time", robust=True)
<xarray.plot.facetgrid.FacetGrid at 0x2b41fcb93970>
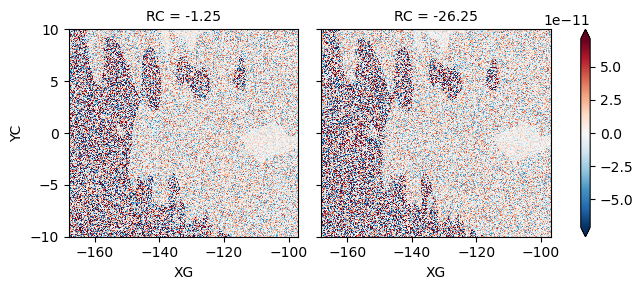
Test : U budget#
RHS = (
budgets.SSHx
+ budgets.Um_dPHdx
+ budgets.Um_Advec
# + budget.Um_Cori # is in Um_Advec
+ budgets.Um_Diss
+ budgets.Um_Impl
+ budgets.Um_Ext.fillna(0)
+ budgets.AB_gU
)
LHS = budgets.TOTUTEND / 86400
(LHS - RHS).isel(time=0, RC=[0, 10]).plot(robust=True, col="RC")
<xarray.plot.facetgrid.FacetGrid at 0x2b4291fe68b0>
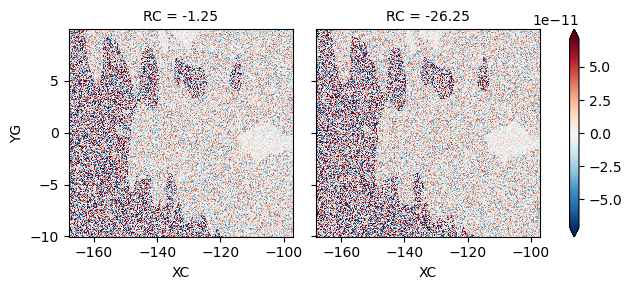
test: V budget#
RHS = (
budgets.SSHy
+ budgets.Vm_dPHdy
+ budgets.Vm_Advec
# + budget.Um_Cori # is in Um_Advec
+ budgets.Vm_Diss
+ budgets.Vm_Impl
+ budgets.Vm_Ext.fillna(0)
+ budgets.AB_gV
)
LHS = budgets.TOTVTEND / 86400
(LHS - RHS).isel(time=0, RC=[0, 10]).plot(robust=True, col="RC")
<xarray.plot.facetgrid.FacetGrid at 0x2b429365b220>
ROMS#
Todo /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_runs_setup_scripts/tpos-DIABLO/diablo_2.0/tpos_config_files/ROMS_PSH_6HRLIN_0N140W_108x108x288_5OCT2020_fixedeps/
Staggered grid
Update grid
something is wrong with your ICs (ocean_ic_S.txt and ocean_ic_T.txt and ocean_ic_U.txt). The values are much too small, except for U = -1 + epsilon. U should be m/s (anomaly). S psu (anomaly). T deg C or Kelvin (anomaly).
except ocean_frc_time.txt. Something is off there with the units/order of magnitude. You need to make sure the time that you want the LES to start is zero, so you have to subtract off the initial time and then have all times as seconds after (and before) that point. I think it is necessary to have one time index (i.e. one negative ocean_frc_time value) before the targeted LES start time for interpolation.
please add the time step before each profile as in ocean_colpsh_T.txt in this directory:/glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_runs_setup_scripts/tpos-DIABLO/diablo_2.0/tpos_config_files
put the outputs in units of deg C per day, m/s per day, psu per day. (I changed the LES to take in units of per day to mitigate precision issues).
You need ocean_colfrc_*.txt files, which are for restoring. They are just the basic fields, T(z,t), U(z,t), S(z,t) and V(z,t) (called “W” in LES world; don’t ask…).
import xroms
outdir = "/glade/work/dcherian/pump/les_forcing/roms_test/"
dirname = "/glade/p/cgd/oce/people/dwhitt/TPOS/tpos20/OUT/"
files = [
f"{dirname}/ocean_dia_tpos20_3_d2_2.nc",
f"{dirname}/ocean_avg_tpos20_3_d2_2.nc",
]
chunk = {"xi": 500, "eta": 120}
ds0 = pump.model.model.read_roms_dataset(files, xi=20, eta=500, ocean_time=50)
rds, grid = xroms.roms_dataset(ds0)
rds["u"] = grid.interp(rds.u, axis="X", boundary="extend")
rds["v"] = grid.interp(rds.v, axis="Y", boundary="extend")
romsds = pump.model.model.make_cartesian(rds)
romsds
/glade/u/home/dcherian/pump/pump/model/model.py:224: RuntimeWarning: Converting a CFTimeIndex with dates from a non-standard calendar, 'noleap', to a pandas.DatetimeIndex, which uses dates from the standard calendar. This may lead to subtle errors in operations that depend on the length of time between dates.
ds["ocean_time"] = ds.indexes["ocean_time"].to_datetimeindex()
<xarray.Dataset>
Dimensions: (boundary: 4, lat_rho: 481, lat_v: 480, lon_rho: 1501, lon_u: 1500, ocean_time: 140, s_rho: 50, s_w: 51, tracer: 2)
Coordinates:
ntimes int32 25201
ndtfast int32 20
dt float64 120.0
dtfast float64 6.0
dstart object 0027-09-02 00:00:00
nHIS int32 3600
ndefHIS int32 0
nRST int32 3600
ntsAVG int32 1
nAVG int32 180
ndefAVG int32 0
ntsDIA int32 1
nDIA int32 180
ndefDIA int32 0
nSTA int32 30
Falpha float64 2.0
Fgamma float64 0.284
nl_tnu4 (tracer) float64 dask.array<chunksize=(2,), meta=np.ndarray>
nl_visc4 float64 1.24e+09
LuvSponge int32 0
LtracerSponge (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
Akt_bak (tracer) float64 dask.array<chunksize=(2,), meta=np.ndarray>
Akv_bak float64 1e-05
rdrg float64 0.000265
rdrg2 float64 0.003
Zob float64 0.02
Zos float64 0.02
Znudg float64 0.0
M2nudg float64 0.0
M3nudg float64 1.0
Tnudg (tracer) float64 dask.array<chunksize=(2,), meta=np.ndarray>
FSobc_in (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
FSobc_out (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
M2obc_in (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
M2obc_out (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
Tobc_in (boundary, tracer) float64 dask.array<chunksize=(4, 2), meta=np.ndarray>
Tobc_out (boundary, tracer) float64 dask.array<chunksize=(4, 2), meta=np.ndarray>
M3obc_in (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
M3obc_out (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
rho0 float64 1.025e+03
gamma2 float64 1.0
LuvSrc int32 0
LwSrc int32 0
LtracerSrc (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
LsshCLM int32 0
Lm2CLM int32 0
Lm3CLM int32 0
LtracerCLM (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
LnudgeM2CLM int32 0
LnudgeM3CLM int32 0
LnudgeTCLM (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
Lm2PSH int32 0
Lm3PSH int32 0
LtracerPSH (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
spherical int32 1
xl float64 8.163e+06
el float64 2.674e+06
Vtransform int32 1
Vstretching int32 1
Tcline float64 75.0
hc float64 75.0
grid int32 1
* s_rho (s_rho) float64 -0.99 -0.97 -0.95 -0.93 ... -0.05 -0.03 -0.01
* s_w (s_w) float64 -1.0 -0.98 -0.96 -0.94 ... -0.04 -0.02 0.0
Cs_r (s_rho) float64 dask.array<chunksize=(50,), meta=np.ndarray>
Cs_w (s_w) float64 dask.array<chunksize=(51,), meta=np.ndarray>
h (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
f (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
pm (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
pn (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
* lon_rho (lon_rho) float64 -170.0 -170.0 -169.9 ... -95.1 -95.05 -95.0
* lat_rho (lat_rho) float64 -12.0 -11.95 -11.9 ... 11.9 11.95 12.0
* lon_u (lon_u) float64 -170.0 -169.9 -169.9 ... -95.12 -95.07 -95.02
lat_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
lon_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
* lat_v (lat_v) float64 -11.97 -11.92 -11.87 ... 11.87 11.92 11.97
lon_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
lat_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
angle (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
mask_rho (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
mask_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
mask_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
mask_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
* ocean_time (ocean_time) datetime64[ns] 1985-10-02T03:00:00 ... 1985-1...
z_w (ocean_time, s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
z_w_u (ocean_time, s_w, lat_rho, lon_u) float64 dask.array<chunksize=(50, 51, 481, 19), meta=np.ndarray>
z_w_v (ocean_time, s_w, lat_v, lon_rho) float64 dask.array<chunksize=(50, 51, 480, 20), meta=np.ndarray>
z_w_psi (ocean_time, s_w, lat_v, lon_u) float64 dask.array<chunksize=(50, 51, 480, 19), meta=np.ndarray>
z_rho (ocean_time, s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
z_rho_u (ocean_time, s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
z_rho_v (ocean_time, s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
z_rho_psi (ocean_time, s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
z_rho0 (s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
z_rho_u0 (s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
z_rho_v0 (s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
z_rho_psi0 (s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
z_w0 (s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(51, 481, 20), meta=np.ndarray>
z_w_u0 (s_w, lat_rho, lon_u) float64 dask.array<chunksize=(51, 481, 19), meta=np.ndarray>
z_w_v0 (s_w, lat_v, lon_rho) float64 dask.array<chunksize=(51, 480, 20), meta=np.ndarray>
z_w_psi0 (s_w, lat_v, lon_u) float64 dask.array<chunksize=(51, 480, 19), meta=np.ndarray>
Dimensions without coordinates: boundary, tracer
Data variables:
zeta (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
ubar (ocean_time, lat_rho, lon_u) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
vbar (ocean_time, lat_v, lon_rho) float32 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
u (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 1), meta=np.ndarray>
v (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 1, 20), meta=np.ndarray>
w (ocean_time, s_w, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
temp (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
Hsbl (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
AKv (ocean_time, s_w, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
AKt (ocean_time, s_w, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
shflux (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
ssflux (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
latent (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
sensible (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
lwrad (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
evaporation (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
rain (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
swrad (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
sustr (ocean_time, lat_rho, lon_u) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
svstr (ocean_time, lat_v, lon_rho) float32 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
u_cor (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_cor (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_vadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_vadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_hadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_hadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_xadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_xadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_yadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_yadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_prsgrd (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_prsgrd (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_vvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_vvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_hvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_hvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_xvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_xvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_yvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_yvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_accel (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_accel (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
temp_hadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_xadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_yadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_vadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_hdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_xdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_ydiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_sdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_vdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_rate (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_hadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_xadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_yadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_vadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_hdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_xdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_ydiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_sdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_vdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_rate (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
pm_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
pn_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
pm_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
pn_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
pm_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
pn_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dx (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
dx_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
dx_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
dx_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dy (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
dy_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
dy_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
dy_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dz (ocean_time, s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
dz_w (ocean_time, s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
dz_u (ocean_time, s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
dz_w_u (ocean_time, s_w, lat_rho, lon_u) float64 dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
dz_v (ocean_time, s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
dz_w_v (ocean_time, s_w, lat_v, lon_rho) float64 dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
dz_psi (ocean_time, s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
dz_w_psi (ocean_time, s_w, lat_v, lon_u) float64 dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
dz0 (s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
dz_w0 (s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(1, 481, 20), meta=np.ndarray>
dz_u0 (s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
dz_w_u0 (s_w, lat_rho, lon_u) float64 dask.array<chunksize=(1, 481, 19), meta=np.ndarray>
dz_v0 (s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
dz_w_v0 (s_w, lat_v, lon_rho) float64 dask.array<chunksize=(1, 480, 20), meta=np.ndarray>
dz_psi0 (s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
dz_w_psi0 (s_w, lat_v, lon_u) float64 dask.array<chunksize=(1, 480, 19), meta=np.ndarray>
dA (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
dA_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
dA_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
dA_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dV (ocean_time, s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
dV_w (ocean_time, s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
dV_u (ocean_time, s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
dV_w_u (ocean_time, s_w, lat_rho, lon_u) float64 dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
dV_v (ocean_time, s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
dV_w_v (ocean_time, s_w, lat_v, lon_rho) float64 dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
dV_psi (ocean_time, s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
dV_w_psi (ocean_time, s_w, lat_v, lon_u) float64 dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
Attributes:
file: /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2...
format: netCDF-3 64bit offset file
Conventions: CF-1.4, SGRID-0.3
type: ROMS/TOMS diagnostics file
title: TPOS 1/20deg
var_info: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
rst_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2...
avg_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2...
dia_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2...
sta_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2...
grd_file: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
ini_file: /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
frc_file_01: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
frc_file_02: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
frc_file_03: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
bry_file: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
script_file: ./tpos20_3_diag_2.in
spos_file: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
NLM_LBC: \nEDGE: WEST SOUTH EAST NORTH \nzeta: Che C...
svn_url: https:://myroms.org/svn/src
svn_rev: Unversioned directory
code_dir: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROM...
header_dir: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
header_file: tpos20.h
os: Linux
cpu: x86_64
compiler_system: ifort
compiler_command: /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/...
compiler_flags: -fp-model precise -ip -O3
tiling: 018x012
history: ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:5...
ana_file: ROMS/Functionals/ana_btflux.h
CPP_options: TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SH...- boundary: 4
- lat_rho: 481
- lat_v: 480
- lon_rho: 1501
- lon_u: 1500
- ocean_time: 140
- s_rho: 50
- s_w: 51
- tracer: 2
- ntimes()int3225201
- long_name :
- number of long time-steps
array(25201, dtype=int32)
- ndtfast()int3220
- long_name :
- number of short time-steps
array(20, dtype=int32)
- dt()float64120.0
- long_name :
- size of long time-steps
- units :
- second
array(120.)
- dtfast()float646.0
- long_name :
- size of short time-steps
- units :
- second
array(6.)
- dstart()object0027-09-02 00:00:00
- long_name :
- time stamp assigned to model initilization
array(cftime.DatetimeNoLeap(27, 9, 2, 0, 0, 0, 0), dtype=object)
- nHIS()int323600
- long_name :
- number of time-steps between history records
array(3600, dtype=int32)
- ndefHIS()int320
- long_name :
- number of time-steps between the creation of history files
array(0, dtype=int32)
- nRST()int323600
- long_name :
- number of time-steps between restart records
- cycle :
- only latest two records are maintained
array(3600, dtype=int32)
- ntsAVG()int321
- long_name :
- starting time-step for accumulation of time-averaged fields
array(1, dtype=int32)
- nAVG()int32180
- long_name :
- number of time-steps between time-averaged records
array(180, dtype=int32)
- ndefAVG()int320
- long_name :
- number of time-steps between the creation of average files
array(0, dtype=int32)
- ntsDIA()int321
- long_name :
- starting time-step for accumulation of diagnostic fields
array(1, dtype=int32)
- nDIA()int32180
- long_name :
- number of time-steps between diagnostic records
array(180, dtype=int32)
- ndefDIA()int320
- long_name :
- number of time-steps between the creation of diagnostic files
array(0, dtype=int32)
- nSTA()int3230
- long_name :
- number of time-steps between stations records
array(30, dtype=int32)
- Falpha()float642.0
- long_name :
- Power-law shape barotropic filter parameter
array(2.)
- Fgamma()float640.284
- long_name :
- Power-law shape barotropic filter parameter
array(0.284)
- nl_tnu4(tracer)float64dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- nonlinear model biharmonic mixing coefficient for tracers
- units :
- meter4 second-1
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - nl_visc4()float641.24e+09
- long_name :
- nonlinear model biharmonic mixing coefficient for momentum
- units :
- meter4 second-1
array(1.24e+09)
- LuvSponge()int320
- long_name :
- horizontal viscosity sponge activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerSponge(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- horizontal diffusivity sponge activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - Akt_bak(tracer)float64dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- background vertical mixing coefficient for tracers
- units :
- meter2 second-1
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Akv_bak()float641e-05
- long_name :
- background vertical mixing coefficient for momentum
- units :
- meter2 second-1
array(1.e-05)
- rdrg()float640.000265
- long_name :
- linear drag coefficient
- units :
- meter second-1
array(0.000265)
- rdrg2()float640.003
- long_name :
- quadratic drag coefficient
array(0.003)
- Zob()float640.02
- long_name :
- bottom roughness
- units :
- meter
array(0.02)
- Zos()float640.02
- long_name :
- surface roughness
- units :
- meter
array(0.02)
- Znudg()float640.0
- long_name :
- free-surface nudging/relaxation inverse time scale
- units :
- day-1
array(0.)
- M2nudg()float640.0
- long_name :
- 2D momentum nudging/relaxation inverse time scale
- units :
- day-1
array(0.)
- M3nudg()float641.0
- long_name :
- 3D momentum nudging/relaxation inverse time scale
- units :
- day-1
array(1.)
- Tnudg(tracer)float64dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- Tracers nudging/relaxation inverse time scale
- units :
- day-1
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - FSobc_in(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- free-surface inflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - FSobc_out(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- free-surface outflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M2obc_in(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 2D momentum inflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M2obc_out(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 2D momentum outflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Tobc_in(boundary, tracer)float64dask.array<chunksize=(4, 2), meta=np.ndarray>
- long_name :
- tracers inflow, nudging inverse time scale
- units :
- second-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 64 B 64 B Shape (4, 2) (4, 2) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Tobc_out(boundary, tracer)float64dask.array<chunksize=(4, 2), meta=np.ndarray>
- long_name :
- tracers outflow, nudging inverse time scale
- units :
- second-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 64 B 64 B Shape (4, 2) (4, 2) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M3obc_in(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 3D momentum inflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M3obc_out(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 3D momentum outflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - rho0()float641.025e+03
- long_name :
- mean density used in Boussinesq approximation
- units :
- kilogram meter-3
array(1025.)
- gamma2()float641.0
- long_name :
- slipperiness parameter
array(1.)
- LuvSrc()int320
- long_name :
- momentum point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LwSrc()int320
- long_name :
- mass point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerSrc(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - LsshCLM()int320
- long_name :
- sea surface height climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Lm2CLM()int320
- long_name :
- 2D momentum climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Lm3CLM()int320
- long_name :
- 3D momentum climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerCLM(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - LnudgeM2CLM()int320
- long_name :
- 2D momentum climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LnudgeM3CLM()int320
- long_name :
- 3D momentum climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LnudgeTCLM(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - Lm2PSH()int320
- long_name :
- 2D momentum push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Lm3PSH()int320
- long_name :
- 3D momentum push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerPSH(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - spherical()int321
- long_name :
- grid type logical switch
- flag_values :
- [0 1]
- flag_meanings :
- Cartesian spherical
array(1, dtype=int32)
- xl()float648.163e+06
- long_name :
- domain length in the XI-direction
- units :
- meter
array(8163221.23397392)
- el()float642.674e+06
- long_name :
- domain length in the ETA-direction
- units :
- meter
array(2674366.462091)
- Vtransform()int321
- long_name :
- vertical terrain-following transformation equation
array(1, dtype=int32)
- Vstretching()int321
- long_name :
- vertical terrain-following stretching function
array(1, dtype=int32)
- Tcline()float6475.0
- long_name :
- S-coordinate surface/bottom layer width
- units :
- meter
array(75.)
- hc()float6475.0
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
array(75.)
- grid()int321
- cf_role :
- grid_topology
- topology_dimension :
- 2
- node_dimensions :
- xi_psi eta_psi
- face_dimensions :
- xi_rho: xi_psi (padding: both) eta_rho: eta_psi (padding: both)
- edge1_dimensions :
- xi_u: xi_psi eta_u: eta_psi (padding: both)
- edge2_dimensions :
- xi_v: xi_psi (padding: both) eta_v: eta_psi
- node_coordinates :
- lon_psi lat_psi
- face_coordinates :
- lon_rho lat_rho
- edge1_coordinates :
- lon_u lat_u
- edge2_coordinates :
- lon_v lat_v
- vertical_dimensions :
- s_rho: s_w (padding: none)
array(1, dtype=int32)
- s_rho(s_rho)float64-0.99 -0.97 -0.95 ... -0.03 -0.01
- long_name :
- S-coordinate at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- standard_name :
- ocean_s_coordinate_g1
- formula_terms :
- s: s_rho C: Cs_r eta: zeta depth: h depth_c: hc
- field :
- s_rho, scalar
- axis :
- Z
array([-0.99, -0.97, -0.95, -0.93, -0.91, -0.89, -0.87, -0.85, -0.83, -0.81, -0.79, -0.77, -0.75, -0.73, -0.71, -0.69, -0.67, -0.65, -0.63, -0.61, -0.59, -0.57, -0.55, -0.53, -0.51, -0.49, -0.47, -0.45, -0.43, -0.41, -0.39, -0.37, -0.35, -0.33, -0.31, -0.29, -0.27, -0.25, -0.23, -0.21, -0.19, -0.17, -0.15, -0.13, -0.11, -0.09, -0.07, -0.05, -0.03, -0.01]) - s_w(s_w)float64-1.0 -0.98 -0.96 ... -0.02 0.0
- long_name :
- S-coordinate at W-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- standard_name :
- ocean_s_coordinate_g1
- formula_terms :
- s: s_w C: Cs_w eta: zeta depth: h depth_c: hc
- field :
- s_w, scalar
- axis :
- Z
array([-1. , -0.98, -0.96, -0.94, -0.92, -0.9 , -0.88, -0.86, -0.84, -0.82, -0.8 , -0.78, -0.76, -0.74, -0.72, -0.7 , -0.68, -0.66, -0.64, -0.62, -0.6 , -0.58, -0.56, -0.54, -0.52, -0.5 , -0.48, -0.46, -0.44, -0.42, -0.4 , -0.38, -0.36, -0.34, -0.32, -0.3 , -0.28, -0.26, -0.24, -0.22, -0.2 , -0.18, -0.16, -0.14, -0.12, -0.1 , -0.08, -0.06, -0.04, -0.02, 0. ]) - Cs_r(s_rho)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- S-coordinate stretching curves at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- field :
- Cs_r, scalar
Array Chunk Bytes 400 B 400 B Shape (50,) (50,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Cs_w(s_w)float64dask.array<chunksize=(51,), meta=np.ndarray>
- long_name :
- S-coordinate stretching curves at W-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- field :
- Cs_w, scalar
Array Chunk Bytes 408 B 408 B Shape (51,) (51,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - h(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- bathymetry at RHO-points
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- bath, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - f(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- Coriolis parameter at RHO-points
- units :
- second-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- coriolis, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - pm(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in XI
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pm, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - pn(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in ETA
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pn, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - lon_rho(lon_rho)float64-170.0 -170.0 ... -95.05 -95.0
- long_name :
- longitude of RHO-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_rho, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- X
array([-170. , -169.95, -169.9 , ..., -95.1 , -95.05, -95. ])
- lat_rho(lat_rho)float64-12.0 -11.95 -11.9 ... 11.95 12.0
- long_name :
- latitude of RHO-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_rho, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- Y
array([-11.999981, -11.949983, -11.899983, ..., 11.899983, 11.949983, 11.999981]) - lon_u(lon_u)float64-170.0 -169.9 ... -95.07 -95.02
- long_name :
- longitude of U-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_u, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- X
array([-169.975, -169.925, -169.875, ..., -95.125, -95.075, -95.025])
- lat_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- latitude of U-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_u, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 76.96 kB Shape (481, 1500) (481, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - lon_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- longitude of V-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_v, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - lat_v(lat_v)float64-11.97 -11.92 ... 11.92 11.97
- long_name :
- latitude of V-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_v, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- Y
array([-11.974982, -11.924983, -11.874983, ..., 11.874983, 11.924983, 11.974982]) - lon_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- longitude of PSI-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_psi, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1500) (480, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - lat_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- latitude of PSI-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_psi, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1500) (480, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - angle(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- angle between XI-axis and EAST
- units :
- radians
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- angle, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - mask_rho(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- mask on RHO-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - mask_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- mask on U-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
Array Chunk Bytes 5.77 MB 76.96 kB Shape (481, 1500) (481, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - mask_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- mask on V-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - mask_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- mask on psi-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- node
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1500) (480, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - ocean_time(ocean_time)datetime64[ns]1985-10-02T03:00:00 ... 1985-11-...
- axis :
- T
- standard_name :
- time
array(['1985-10-02T03:00:00.000000000', '1985-10-02T09:00:00.000000000', '1985-10-02T15:00:00.000000000', '1985-10-02T21:00:00.000000000', '1985-10-03T03:00:00.000000000', '1985-10-03T09:00:00.000000000', '1985-10-03T15:00:00.000000000', '1985-10-03T21:00:00.000000000', '1985-10-04T03:00:00.000000000', '1985-10-04T09:00:00.000000000', '1985-10-04T15:00:00.000000000', '1985-10-04T21:00:00.000000000', '1985-10-05T03:00:00.000000000', '1985-10-05T09:00:00.000000000', '1985-10-05T15:00:00.000000000', '1985-10-05T21:00:00.000000000', '1985-10-06T03:00:00.000000000', '1985-10-06T09:00:00.000000000', '1985-10-06T15:00:00.000000000', '1985-10-06T21:00:00.000000000', '1985-10-07T03:00:00.000000000', '1985-10-07T09:00:00.000000000', '1985-10-07T15:00:00.000000000', '1985-10-07T21:00:00.000000000', '1985-10-08T03:00:00.000000000', '1985-10-08T09:00:00.000000000', '1985-10-08T15:00:00.000000000', '1985-10-08T21:00:00.000000000', '1985-10-09T03:00:00.000000000', '1985-10-09T09:00:00.000000000', '1985-10-09T15:00:00.000000000', '1985-10-09T21:00:00.000000000', '1985-10-10T03:00:00.000000000', '1985-10-10T09:00:00.000000000', '1985-10-10T15:00:00.000000000', '1985-10-10T21:00:00.000000000', '1985-10-11T03:00:00.000000000', '1985-10-11T09:00:00.000000000', '1985-10-11T15:00:00.000000000', '1985-10-11T21:00:00.000000000', '1985-10-12T03:00:00.000000000', '1985-10-12T09:00:00.000000000', '1985-10-12T15:00:00.000000000', '1985-10-12T21:00:00.000000000', '1985-10-13T03:00:00.000000000', '1985-10-13T09:00:00.000000000', '1985-10-13T15:00:00.000000000', '1985-10-13T21:00:00.000000000', '1985-10-14T03:00:00.000000000', '1985-10-14T09:00:00.000000000', '1985-10-14T15:00:00.000000000', '1985-10-14T21:00:00.000000000', '1985-10-15T03:00:00.000000000', '1985-10-15T09:00:00.000000000', '1985-10-15T15:00:00.000000000', '1985-10-15T21:00:00.000000000', '1985-10-16T03:00:00.000000000', '1985-10-16T09:00:00.000000000', '1985-10-16T15:00:00.000000000', '1985-10-16T21:00:00.000000000', '1985-10-17T03:00:00.000000000', '1985-10-17T09:00:00.000000000', '1985-10-17T15:00:00.000000000', '1985-10-17T21:00:00.000000000', '1985-10-18T03:00:00.000000000', '1985-10-18T09:00:00.000000000', '1985-10-18T15:00:00.000000000', '1985-10-18T21:00:00.000000000', '1985-10-19T03:00:00.000000000', '1985-10-19T09:00:00.000000000', '1985-10-19T15:00:00.000000000', '1985-10-19T21:00:00.000000000', '1985-10-20T03:00:00.000000000', '1985-10-20T09:00:00.000000000', '1985-10-20T15:00:00.000000000', '1985-10-20T21:00:00.000000000', '1985-10-21T03:00:00.000000000', '1985-10-21T09:00:00.000000000', '1985-10-21T15:00:00.000000000', '1985-10-21T21:00:00.000000000', '1985-10-22T03:00:00.000000000', '1985-10-22T09:00:00.000000000', '1985-10-22T15:00:00.000000000', '1985-10-22T21:00:00.000000000', '1985-10-23T03:00:00.000000000', '1985-10-23T09:00:00.000000000', '1985-10-23T15:00:00.000000000', '1985-10-23T21:00:00.000000000', '1985-10-24T03:00:00.000000000', '1985-10-24T09:00:00.000000000', '1985-10-24T15:00:00.000000000', '1985-10-24T21:00:00.000000000', '1985-10-25T03:00:00.000000000', '1985-10-25T09:00:00.000000000', '1985-10-25T15:00:00.000000000', '1985-10-25T21:00:00.000000000', '1985-10-26T03:00:00.000000000', '1985-10-26T09:00:00.000000000', '1985-10-26T15:00:00.000000000', '1985-10-26T21:00:00.000000000', '1985-10-27T03:00:00.000000000', '1985-10-27T09:00:00.000000000', '1985-10-27T15:00:00.000000000', '1985-10-27T21:00:00.000000000', '1985-10-28T03:00:00.000000000', '1985-10-28T09:00:00.000000000', '1985-10-28T15:00:00.000000000', '1985-10-28T21:00:00.000000000', '1985-10-29T03:00:00.000000000', '1985-10-29T09:00:00.000000000', '1985-10-29T15:00:00.000000000', '1985-10-29T21:00:00.000000000', '1985-10-30T03:00:00.000000000', '1985-10-30T09:00:00.000000000', '1985-10-30T15:00:00.000000000', '1985-10-30T21:00:00.000000000', '1985-10-31T03:00:00.000000000', '1985-10-31T09:00:00.000000000', '1985-10-31T15:00:00.000000000', '1985-10-31T21:00:00.000000000', '1985-11-01T03:00:00.000000000', '1985-11-01T09:00:00.000000000', '1985-11-01T15:00:00.000000000', '1985-11-01T21:00:00.000000000', '1985-11-02T03:00:00.000000000', '1985-11-02T09:00:00.000000000', '1985-11-02T15:00:00.000000000', '1985-11-02T21:00:00.000000000', '1985-11-03T03:00:00.000000000', '1985-11-03T09:00:00.000000000', '1985-11-03T15:00:00.000000000', '1985-11-03T21:00:00.000000000', '1985-11-04T03:00:00.000000000', '1985-11-04T09:00:00.000000000', '1985-11-04T15:00:00.000000000', '1985-11-04T21:00:00.000000000', '1985-11-05T03:00:00.000000000', '1985-11-05T09:00:00.000000000', '1985-11-05T15:00:00.000000000', '1985-11-05T21:00:00.000000000'], dtype='datetime64[ns]') - z_w(ocean_time, s_w, lat_rho, lon_rho)float64dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.24 GB 196.25 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 2368 Tasks 228 Chunks Type float64 numpy.ndarray - z_w_u(ocean_time, s_w, lat_rho, lon_u)float64dask.array<chunksize=(50, 51, 481, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.21 GB 186.44 MB Shape (140, 51, 481, 1500) (50, 51, 481, 19) Count 5515 Tasks 450 Chunks Type float64 numpy.ndarray - z_w_v(ocean_time, s_w, lat_v, lon_rho)float64dask.array<chunksize=(50, 51, 480, 20), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.15 GB 195.84 MB Shape (140, 51, 480, 1501) (50, 51, 480, 20) Count 3280 Tasks 228 Chunks Type float64 numpy.ndarray - z_w_psi(ocean_time, s_w, lat_v, lon_u)float64dask.array<chunksize=(50, 51, 480, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.13 GB 186.05 MB Shape (140, 51, 480, 1500) (50, 51, 480, 19) Count 7315 Tasks 450 Chunks Type float64 numpy.ndarray - z_rho(ocean_time, s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.43 GB 192.40 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 2368 Tasks 228 Chunks Type float64 numpy.ndarray - z_rho_u(ocean_time, s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.40 GB 182.78 MB Shape (140, 50, 481, 1500) (50, 50, 481, 19) Count 5515 Tasks 450 Chunks Type float64 numpy.ndarray - z_rho_v(ocean_time, s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.35 GB 192.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 3280 Tasks 228 Chunks Type float64 numpy.ndarray - z_rho_psi(ocean_time, s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.32 GB 182.40 MB Shape (140, 50, 480, 1500) (50, 50, 480, 19) Count 7315 Tasks 450 Chunks Type float64 numpy.ndarray - z_rho0(s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.79 MB 3.85 MB Shape (50, 481, 1501) (50, 481, 20) Count 315 Tasks 76 Chunks Type float64 numpy.ndarray - z_rho_u0(s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.60 MB 3.66 MB Shape (50, 481, 1500) (50, 481, 19) Count 1364 Tasks 150 Chunks Type float64 numpy.ndarray - z_rho_v0(s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.19 MB 3.84 MB Shape (50, 480, 1501) (50, 480, 20) Count 619 Tasks 76 Chunks Type float64 numpy.ndarray - z_rho_psi0(s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.00 MB 3.65 MB Shape (50, 480, 1500) (50, 480, 19) Count 1964 Tasks 150 Chunks Type float64 numpy.ndarray - z_w0(s_w, lat_rho, lon_rho)float64dask.array<chunksize=(51, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.57 MB 3.92 MB Shape (51, 481, 1501) (51, 481, 20) Count 315 Tasks 76 Chunks Type float64 numpy.ndarray - z_w_u0(s_w, lat_rho, lon_u)float64dask.array<chunksize=(51, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.37 MB 3.73 MB Shape (51, 481, 1500) (51, 481, 19) Count 1364 Tasks 150 Chunks Type float64 numpy.ndarray - z_w_v0(s_w, lat_v, lon_rho)float64dask.array<chunksize=(51, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.96 MB 3.92 MB Shape (51, 480, 1501) (51, 480, 20) Count 619 Tasks 76 Chunks Type float64 numpy.ndarray - z_w_psi0(s_w, lat_v, lon_u)float64dask.array<chunksize=(51, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.76 MB 3.72 MB Shape (51, 480, 1500) (51, 480, 19) Count 1964 Tasks 150 Chunks Type float64 numpy.ndarray
- zeta(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged free-surface
- units :
- meter
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- free-surface, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - ubar(ocean_time, lat_rho, lon_u)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged vertically integrated u-momentum component
- units :
- meter second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- ubar-velocity, scalar, series
Array Chunk Bytes 404.04 MB 1.92 MB Shape (140, 481, 1500) (50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - vbar(ocean_time, lat_v, lon_rho)float32dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- long_name :
- time-averaged vertically integrated v-momentum component
- units :
- meter second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- vbar-velocity, scalar, series
Array Chunk Bytes 403.47 MB 1.92 MB Shape (140, 480, 1501) (50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 1), meta=np.ndarray>
Array Chunk Bytes 20.22 GB 91.39 MB Shape (140, 50, 481, 1501) (50, 50, 481, 19) Count 3400 Tasks 453 Chunks Type float32 numpy.ndarray - v(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 1, 20), meta=np.ndarray>
Array Chunk Bytes 20.22 GB 95.80 MB Shape (140, 50, 481, 1501) (50, 50, 479, 20) Count 5245 Tasks 684 Chunks Type float32 numpy.ndarray - w(ocean_time, s_w, lat_rho, lon_rho)float32dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged vertical momentum component
- units :
- meter second-1
- time :
- ocean_time
- standard_name :
- upward_sea_water_velocity
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- w-velocity, scalar, series
Array Chunk Bytes 20.62 GB 98.12 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged potential temperature
- units :
- Celsius
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temperature, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged salinity
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salinity, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - Hsbl(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged depth of oceanic surface boundary layer
- units :
- meter
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- Hsbl, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - AKv(ocean_time, s_w, lat_rho, lon_rho)float32dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged vertical viscosity coefficient
- units :
- meter2 second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- AKv, scalar, series
Array Chunk Bytes 20.62 GB 98.12 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - AKt(ocean_time, s_w, lat_rho, lon_rho)float32dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged temperature vertical diffusion coefficient
- units :
- meter2 second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- AKt, scalar, series
Array Chunk Bytes 20.62 GB 98.12 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - shflux(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged surface net heat flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- surface heat flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - ssflux(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged surface net salt flux, (E-P)*SALT
- units :
- meter second-1
- negative_value :
- upward flux, freshening (net precipitation)
- positive_value :
- downward flux, salting (net evaporation)
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- surface net salt flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - latent(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged net latent heat flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- latent heat flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - sensible(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged net sensible heat flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- sensible heat flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - lwrad(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged net longwave radiation flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- longwave radiation, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - evaporation(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged evaporation rate
- units :
- kilogram meter-2 second-1
- negative_value :
- downward flux, freshening (condensation)
- positive_value :
- upward flux, salting (evaporation)
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- evaporation, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - rain(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged rain fall rate
- units :
- kilogram meter-2 second-1
- positive_value :
- downward flux, freshening (precipitation)
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- rain, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - swrad(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged solar shortwave radiation flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- shortwave radiation, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - sustr(ocean_time, lat_rho, lon_u)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged surface u-momentum stress
- units :
- newton meter-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- surface u-momentum stress, scalar, series
Array Chunk Bytes 404.04 MB 1.92 MB Shape (140, 481, 1500) (50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - svstr(ocean_time, lat_v, lon_rho)float32dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- long_name :
- time-averaged surface v-momentum stress
- units :
- newton meter-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- surface v-momentum stress, scalar, series
Array Chunk Bytes 403.47 MB 1.92 MB Shape (140, 480, 1501) (50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_cor(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, Coriolis term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_cor, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_cor(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, Coriolis term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_cor, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_vadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, vertical advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_vadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_vadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, vertical advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_vadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_hadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_hadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_hadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_hadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_xadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal XI-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_xadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_xadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal XI-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_xadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_yadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal ETA-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_yadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_yadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal ETA-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_yadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_prsgrd(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, pressure gradient term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_prsgrd, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_prsgrd(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, pressure gradient term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_prsgrd, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_vvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, vertical viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_vvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_vvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, vertical viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_vvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_hvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_hvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_hvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_hvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_xvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal XI-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_xvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_xvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal XI-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_xvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_yvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal ETA-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_yvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_yvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal ETA-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_yvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_accel(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, acceleration term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_accel, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_accel(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, acceleration term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_accel, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_hadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_hadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_xadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal XI-advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_xadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_yadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal ETA-advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_yadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_vadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, vertical advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_vadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_hdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_hdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_xdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal XI-diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_xdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_ydiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal ETA-diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_ydiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_sdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal S-diffusion rotated tensor term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_sdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_vdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, vertical diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_vdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_rate(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, time rate of change
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_rate, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_hadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_hadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_xadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal XI-advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_xadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_yadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal ETA-advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_yadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_vadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, vertical advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_vadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_hdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_hdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_xdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal XI-diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_xdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_ydiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal ETA-diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_ydiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_sdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal S-diffusion rotated tensor term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_sdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_vdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, vertical diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_vdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_rate(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, time rate of change
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_rate, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - pm_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 381 Tasks 76 Chunks Type float64 numpy.ndarray - pn_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1126 Tasks 150 Chunks Type float64 numpy.ndarray - pm_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1126 Tasks 150 Chunks Type float64 numpy.ndarray - pn_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 381 Tasks 76 Chunks Type float64 numpy.ndarray - pm_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1430 Tasks 150 Chunks Type float64 numpy.ndarray - pn_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1726 Tasks 150 Chunks Type float64 numpy.ndarray - dx(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in XI
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pm, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 153 Tasks 76 Chunks Type float64 numpy.ndarray - dx_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1276 Tasks 150 Chunks Type float64 numpy.ndarray - dx_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 457 Tasks 76 Chunks Type float64 numpy.ndarray - dx_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1580 Tasks 150 Chunks Type float64 numpy.ndarray - dy(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in ETA
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pn, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 153 Tasks 76 Chunks Type float64 numpy.ndarray - dy_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1276 Tasks 150 Chunks Type float64 numpy.ndarray - dy_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 457 Tasks 76 Chunks Type float64 numpy.ndarray - dy_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1876 Tasks 150 Chunks Type float64 numpy.ndarray - dz(ocean_time, s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.43 GB 192.40 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 3052 Tasks 228 Chunks Type float64 numpy.ndarray - dz_w(ocean_time, s_w, lat_rho, lon_rho)float64dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.24 GB 188.55 MB Shape (140, 51, 481, 1501) (50, 49, 481, 20) Count 6472 Tasks 684 Chunks Type float64 numpy.ndarray - dz_u(ocean_time, s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.40 GB 182.78 MB Shape (140, 50, 481, 1500) (50, 50, 481, 19) Count 6199 Tasks 450 Chunks Type float64 numpy.ndarray - dz_w_u(ocean_time, s_w, lat_rho, lon_u)float64dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.21 GB 179.12 MB Shape (140, 51, 481, 1500) (50, 49, 481, 19) Count 15913 Tasks 1350 Chunks Type float64 numpy.ndarray - dz_v(ocean_time, s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.35 GB 192.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 3964 Tasks 228 Chunks Type float64 numpy.ndarray - dz_w_v(ocean_time, s_w, lat_v, lon_rho)float64dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.15 GB 188.16 MB Shape (140, 51, 480, 1501) (50, 49, 480, 20) Count 9208 Tasks 684 Chunks Type float64 numpy.ndarray - dz_psi(ocean_time, s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.32 GB 182.40 MB Shape (140, 50, 480, 1500) (50, 50, 480, 19) Count 7111 Tasks 450 Chunks Type float64 numpy.ndarray - dz_w_psi(ocean_time, s_w, lat_v, lon_u)float64dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.13 GB 178.75 MB Shape (140, 51, 480, 1500) (50, 49, 480, 19) Count 18649 Tasks 1350 Chunks Type float64 numpy.ndarray - dz0(s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.79 MB 3.85 MB Shape (50, 481, 1501) (50, 481, 20) Count 543 Tasks 76 Chunks Type float64 numpy.ndarray - dz_w0(s_w, lat_rho, lon_rho)float64dask.array<chunksize=(1, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.57 MB 3.77 MB Shape (51, 481, 1501) (49, 481, 20) Count 1683 Tasks 228 Chunks Type float64 numpy.ndarray - dz_u0(s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.60 MB 3.66 MB Shape (50, 481, 1500) (50, 481, 19) Count 1592 Tasks 150 Chunks Type float64 numpy.ndarray - dz_w_u0(s_w, lat_rho, lon_u)float64dask.array<chunksize=(1, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.37 MB 3.58 MB Shape (51, 481, 1500) (49, 481, 19) Count 4830 Tasks 450 Chunks Type float64 numpy.ndarray - dz_v0(s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.19 MB 3.84 MB Shape (50, 480, 1501) (50, 480, 20) Count 847 Tasks 76 Chunks Type float64 numpy.ndarray - dz_w_v0(s_w, lat_v, lon_rho)float64dask.array<chunksize=(1, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.96 MB 3.76 MB Shape (51, 480, 1501) (49, 480, 20) Count 2595 Tasks 228 Chunks Type float64 numpy.ndarray - dz_psi0(s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.00 MB 3.65 MB Shape (50, 480, 1500) (50, 480, 19) Count 1896 Tasks 150 Chunks Type float64 numpy.ndarray - dz_w_psi0(s_w, lat_v, lon_u)float64dask.array<chunksize=(1, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.76 MB 3.58 MB Shape (51, 480, 1500) (49, 480, 19) Count 5742 Tasks 450 Chunks Type float64 numpy.ndarray - dA(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in XI
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pm, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 382 Tasks 76 Chunks Type float64 numpy.ndarray - dA_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 2702 Tasks 150 Chunks Type float64 numpy.ndarray - dA_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 990 Tasks 76 Chunks Type float64 numpy.ndarray - dA_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 3606 Tasks 150 Chunks Type float64 numpy.ndarray - dV(ocean_time, s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.43 GB 192.40 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 3966 Tasks 228 Chunks Type float64 numpy.ndarray - dV_w(ocean_time, s_w, lat_rho, lon_rho)float64dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.24 GB 188.55 MB Shape (140, 51, 481, 1501) (50, 49, 481, 20) Count 8298 Tasks 684 Chunks Type float64 numpy.ndarray - dV_u(ocean_time, s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.40 GB 182.78 MB Shape (140, 50, 481, 1500) (50, 50, 481, 19) Count 9951 Tasks 450 Chunks Type float64 numpy.ndarray - dV_w_u(ocean_time, s_w, lat_rho, lon_u)float64dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.21 GB 179.12 MB Shape (140, 51, 481, 1500) (50, 49, 481, 19) Count 21465 Tasks 1350 Chunks Type float64 numpy.ndarray - dV_v(ocean_time, s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.35 GB 192.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 5486 Tasks 228 Chunks Type float64 numpy.ndarray - dV_w_v(ocean_time, s_w, lat_v, lon_rho)float64dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.15 GB 188.16 MB Shape (140, 51, 480, 1501) (50, 49, 480, 20) Count 11642 Tasks 684 Chunks Type float64 numpy.ndarray - dV_psi(ocean_time, s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.32 GB 182.40 MB Shape (140, 50, 480, 1500) (50, 50, 480, 19) Count 11767 Tasks 450 Chunks Type float64 numpy.ndarray - dV_w_psi(ocean_time, s_w, lat_v, lon_u)float64dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.13 GB 178.75 MB Shape (140, 51, 480, 1500) (50, 49, 480, 19) Count 25105 Tasks 1350 Chunks Type float64 numpy.ndarray
- file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- format :
- netCDF-3 64bit offset file
- Conventions :
- CF-1.4, SGRID-0.3
- type :
- ROMS/TOMS diagnostics file
- title :
- TPOS 1/20deg
- var_info :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/varinfo.dat
- rst_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2_2.nc
- avg_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2_2.nc
- dia_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- sta_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2_2.nc
- grd_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_grd.nc
- ini_file :
- /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
- frc_file_01 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_ncep.nc
- frc_file_02 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_giss.nc
- frc_file_03 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_gcgcs.nc
- bry_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_bry.nc
- script_file :
- ./tpos20_3_diag_2.in
- spos_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/stations_tpos20.in
- NLM_LBC :
- EDGE: WEST SOUTH EAST NORTH zeta: Che Che Che Che ubar: Shc Shc Shc Shc vbar: Shc Shc Shc Shc u: RadNud RadNud RadNud RadNud v: RadNud RadNud RadNud RadNud temp: RadNud RadNud Cla RadNud salt: RadNud RadNud Cla RadNud
- svn_url :
- https:://myroms.org/svn/src
- svn_rev :
- Unversioned directory
- code_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROMS/ROMS
- header_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INC/
- header_file :
- tpos20.h
- os :
- Linux
- cpu :
- x86_64
- compiler_system :
- ifort
- compiler_command :
- /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/mpi/mpif90
- compiler_flags :
- -fp-model precise -ip -O3
- tiling :
- 018x012
- history :
- ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:53:43 PM
- ana_file :
- ROMS/Functionals/ana_btflux.h
- CPP_options :
- TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SHAPE, AVERAGES, !BOUNDARY_A BULK_FLUXES, CCSM_FLUXES, !COLLECT_ALL..., CURVGRID, DIAGNOSTICS_TS, DIAGNOSTICS_UV, DJ_GRADPS, DOUBLE_PRECISION, EMINUSP, INLINE_2DIO, LMD_CONVEC, LMD_DDMIX, LMD_MIXING, LMD_NONLOCAL, LMD_RIMIX, LMD_SHAPIRO, LMD_SKPP, LONGWAVE_OUT, MASKING, MIX_ISO_TS, MIX_S_UV, MPI, NONLINEAR, NONLIN_EOS, NO_LBC_ATT, PERFECT_RESTART, POWER_LAW, PROFILE, RADIATION_2D, REDUCE_ALLGATHER, RI_SPLINES, !RST_SINGLE, SALINITY, SOLAR_SOURCE, SOLVE3D, SPLINES_VDIFF, SPLINES_VVISC, STATIONS, TS_U3HADVECTION, TS_C4VADVECTION, TS_DIF4, UV_ADV, UV_COR, UV_U3HADVECTION, UV_C4VADVECTION, UV_QDRAG, UV_VIS4, VAR_RHO_2D
romsds
<xarray.Dataset>
Dimensions: (boundary: 4, lat_rho: 481, lat_v: 480, lon_rho: 1501, lon_u: 1500, ocean_time: 140, s_rho: 50, s_w: 51, tracer: 2)
Coordinates:
ntimes int32 25201
ndtfast int32 20
dt float64 120.0
dtfast float64 6.0
dstart object 0027-09-02 00:00:00
nHIS int32 3600
ndefHIS int32 0
nRST int32 3600
ntsAVG int32 1
nAVG int32 180
ndefAVG int32 0
ntsDIA int32 1
nDIA int32 180
ndefDIA int32 0
nSTA int32 30
Falpha float64 2.0
Fgamma float64 0.284
nl_tnu4 (tracer) float64 dask.array<chunksize=(2,), meta=np.ndarray>
nl_visc4 float64 1.24e+09
LuvSponge int32 0
LtracerSponge (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
Akt_bak (tracer) float64 dask.array<chunksize=(2,), meta=np.ndarray>
Akv_bak float64 1e-05
rdrg float64 0.000265
rdrg2 float64 0.003
Zob float64 0.02
Zos float64 0.02
Znudg float64 0.0
M2nudg float64 0.0
M3nudg float64 1.0
Tnudg (tracer) float64 dask.array<chunksize=(2,), meta=np.ndarray>
FSobc_in (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
FSobc_out (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
M2obc_in (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
M2obc_out (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
Tobc_in (boundary, tracer) float64 dask.array<chunksize=(4, 2), meta=np.ndarray>
Tobc_out (boundary, tracer) float64 dask.array<chunksize=(4, 2), meta=np.ndarray>
M3obc_in (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
M3obc_out (boundary) float64 dask.array<chunksize=(4,), meta=np.ndarray>
rho0 float64 1.025e+03
gamma2 float64 1.0
LuvSrc int32 0
LwSrc int32 0
LtracerSrc (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
LsshCLM int32 0
Lm2CLM int32 0
Lm3CLM int32 0
LtracerCLM (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
LnudgeM2CLM int32 0
LnudgeM3CLM int32 0
LnudgeTCLM (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
Lm2PSH int32 0
Lm3PSH int32 0
LtracerPSH (tracer) int32 dask.array<chunksize=(2,), meta=np.ndarray>
spherical int32 1
xl float64 8.163e+06
el float64 2.674e+06
Vtransform int32 1
Vstretching int32 1
Tcline float64 75.0
hc float64 75.0
grid int32 1
* s_rho (s_rho) float64 -0.99 -0.97 -0.95 -0.93 ... -0.05 -0.03 -0.01
* s_w (s_w) float64 -1.0 -0.98 -0.96 -0.94 ... -0.04 -0.02 0.0
Cs_r (s_rho) float64 dask.array<chunksize=(50,), meta=np.ndarray>
Cs_w (s_w) float64 dask.array<chunksize=(51,), meta=np.ndarray>
h (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
f (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
pm (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
pn (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
* lon_rho (lon_rho) float64 -170.0 -170.0 -169.9 ... -95.1 -95.05 -95.0
* lat_rho (lat_rho) float64 -12.0 -11.95 -11.9 ... 11.9 11.95 12.0
* lon_u (lon_u) float64 -170.0 -169.9 -169.9 ... -95.12 -95.07 -95.02
lat_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
lon_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
* lat_v (lat_v) float64 -11.97 -11.92 -11.87 ... 11.87 11.92 11.97
lon_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
lat_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
angle (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
mask_rho (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
mask_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
mask_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
mask_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
* ocean_time (ocean_time) datetime64[ns] 1985-10-02T03:00:00 ... 1985-1...
z_w (ocean_time, s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
z_w_u (ocean_time, s_w, lat_rho, lon_u) float64 dask.array<chunksize=(50, 51, 481, 19), meta=np.ndarray>
z_w_v (ocean_time, s_w, lat_v, lon_rho) float64 dask.array<chunksize=(50, 51, 480, 20), meta=np.ndarray>
z_w_psi (ocean_time, s_w, lat_v, lon_u) float64 dask.array<chunksize=(50, 51, 480, 19), meta=np.ndarray>
z_rho (ocean_time, s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
z_rho_u (ocean_time, s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
z_rho_v (ocean_time, s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
z_rho_psi (ocean_time, s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
z_rho0 (s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
z_rho_u0 (s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
z_rho_v0 (s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
z_rho_psi0 (s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
z_w0 (s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(51, 481, 20), meta=np.ndarray>
z_w_u0 (s_w, lat_rho, lon_u) float64 dask.array<chunksize=(51, 481, 19), meta=np.ndarray>
z_w_v0 (s_w, lat_v, lon_rho) float64 dask.array<chunksize=(51, 480, 20), meta=np.ndarray>
z_w_psi0 (s_w, lat_v, lon_u) float64 dask.array<chunksize=(51, 480, 19), meta=np.ndarray>
Dimensions without coordinates: boundary, tracer
Data variables:
zeta (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
ubar (ocean_time, lat_rho, lon_u) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
vbar (ocean_time, lat_v, lon_rho) float32 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
u (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 1), meta=np.ndarray>
v (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 1, 20), meta=np.ndarray>
w (ocean_time, s_w, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
temp (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
Hsbl (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
AKv (ocean_time, s_w, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
AKt (ocean_time, s_w, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
shflux (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
ssflux (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
latent (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
sensible (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
lwrad (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
evaporation (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
rain (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
swrad (ocean_time, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
sustr (ocean_time, lat_rho, lon_u) float32 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
svstr (ocean_time, lat_v, lon_rho) float32 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
u_cor (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_cor (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_vadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_vadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_hadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_hadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_xadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_xadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_yadv (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_yadv (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_prsgrd (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_prsgrd (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_vvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_vvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_hvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_hvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_xvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_xvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_yvisc (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_yvisc (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
u_accel (ocean_time, s_rho, lat_rho, lon_u) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
v_accel (ocean_time, s_rho, lat_v, lon_rho) float32 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
temp_hadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_xadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_yadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_vadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_hdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_xdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_ydiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_sdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_vdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
temp_rate (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_hadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_xadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_yadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_vadv (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_hdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_xdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_ydiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_sdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_vdiff (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
salt_rate (ocean_time, s_rho, lat_rho, lon_rho) float32 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
pm_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
pn_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
pm_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
pn_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
pm_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
pn_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dx (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
dx_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
dx_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
dx_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dy (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
dy_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
dy_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
dy_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dz (ocean_time, s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
dz_w (ocean_time, s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
dz_u (ocean_time, s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
dz_w_u (ocean_time, s_w, lat_rho, lon_u) float64 dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
dz_v (ocean_time, s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
dz_w_v (ocean_time, s_w, lat_v, lon_rho) float64 dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
dz_psi (ocean_time, s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
dz_w_psi (ocean_time, s_w, lat_v, lon_u) float64 dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
dz0 (s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
dz_w0 (s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(1, 481, 20), meta=np.ndarray>
dz_u0 (s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
dz_w_u0 (s_w, lat_rho, lon_u) float64 dask.array<chunksize=(1, 481, 19), meta=np.ndarray>
dz_v0 (s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
dz_w_v0 (s_w, lat_v, lon_rho) float64 dask.array<chunksize=(1, 480, 20), meta=np.ndarray>
dz_psi0 (s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
dz_w_psi0 (s_w, lat_v, lon_u) float64 dask.array<chunksize=(1, 480, 19), meta=np.ndarray>
dA (lat_rho, lon_rho) float64 dask.array<chunksize=(481, 20), meta=np.ndarray>
dA_u (lat_rho, lon_u) float64 dask.array<chunksize=(481, 19), meta=np.ndarray>
dA_v (lat_v, lon_rho) float64 dask.array<chunksize=(480, 20), meta=np.ndarray>
dA_psi (lat_v, lon_u) float64 dask.array<chunksize=(480, 19), meta=np.ndarray>
dV (ocean_time, s_rho, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
dV_w (ocean_time, s_w, lat_rho, lon_rho) float64 dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
dV_u (ocean_time, s_rho, lat_rho, lon_u) float64 dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
dV_w_u (ocean_time, s_w, lat_rho, lon_u) float64 dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
dV_v (ocean_time, s_rho, lat_v, lon_rho) float64 dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
dV_w_v (ocean_time, s_w, lat_v, lon_rho) float64 dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
dV_psi (ocean_time, s_rho, lat_v, lon_u) float64 dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
dV_w_psi (ocean_time, s_w, lat_v, lon_u) float64 dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
Attributes:
file: /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2...
format: netCDF-3 64bit offset file
Conventions: CF-1.4, SGRID-0.3
type: ROMS/TOMS diagnostics file
title: TPOS 1/20deg
var_info: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
rst_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2...
avg_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2...
dia_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2...
sta_file: /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2...
grd_file: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
ini_file: /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
frc_file_01: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
frc_file_02: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
frc_file_03: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
bry_file: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
script_file: ./tpos20_3_diag_2.in
spos_file: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
NLM_LBC: \nEDGE: WEST SOUTH EAST NORTH \nzeta: Che C...
svn_url: https:://myroms.org/svn/src
svn_rev: Unversioned directory
code_dir: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROM...
header_dir: /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpo...
header_file: tpos20.h
os: Linux
cpu: x86_64
compiler_system: ifort
compiler_command: /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/...
compiler_flags: -fp-model precise -ip -O3
tiling: 018x012
history: ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:5...
ana_file: ROMS/Functionals/ana_btflux.h
CPP_options: TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SH...- boundary: 4
- lat_rho: 481
- lat_v: 480
- lon_rho: 1501
- lon_u: 1500
- ocean_time: 140
- s_rho: 50
- s_w: 51
- tracer: 2
- ntimes()int3225201
- long_name :
- number of long time-steps
array(25201, dtype=int32)
- ndtfast()int3220
- long_name :
- number of short time-steps
array(20, dtype=int32)
- dt()float64120.0
- long_name :
- size of long time-steps
- units :
- second
array(120.)
- dtfast()float646.0
- long_name :
- size of short time-steps
- units :
- second
array(6.)
- dstart()object0027-09-02 00:00:00
- long_name :
- time stamp assigned to model initilization
array(cftime.DatetimeNoLeap(27, 9, 2, 0, 0, 0, 0), dtype=object)
- nHIS()int323600
- long_name :
- number of time-steps between history records
array(3600, dtype=int32)
- ndefHIS()int320
- long_name :
- number of time-steps between the creation of history files
array(0, dtype=int32)
- nRST()int323600
- long_name :
- number of time-steps between restart records
- cycle :
- only latest two records are maintained
array(3600, dtype=int32)
- ntsAVG()int321
- long_name :
- starting time-step for accumulation of time-averaged fields
array(1, dtype=int32)
- nAVG()int32180
- long_name :
- number of time-steps between time-averaged records
array(180, dtype=int32)
- ndefAVG()int320
- long_name :
- number of time-steps between the creation of average files
array(0, dtype=int32)
- ntsDIA()int321
- long_name :
- starting time-step for accumulation of diagnostic fields
array(1, dtype=int32)
- nDIA()int32180
- long_name :
- number of time-steps between diagnostic records
array(180, dtype=int32)
- ndefDIA()int320
- long_name :
- number of time-steps between the creation of diagnostic files
array(0, dtype=int32)
- nSTA()int3230
- long_name :
- number of time-steps between stations records
array(30, dtype=int32)
- Falpha()float642.0
- long_name :
- Power-law shape barotropic filter parameter
array(2.)
- Fgamma()float640.284
- long_name :
- Power-law shape barotropic filter parameter
array(0.284)
- nl_tnu4(tracer)float64dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- nonlinear model biharmonic mixing coefficient for tracers
- units :
- meter4 second-1
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - nl_visc4()float641.24e+09
- long_name :
- nonlinear model biharmonic mixing coefficient for momentum
- units :
- meter4 second-1
array(1.24e+09)
- LuvSponge()int320
- long_name :
- horizontal viscosity sponge activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerSponge(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- horizontal diffusivity sponge activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - Akt_bak(tracer)float64dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- background vertical mixing coefficient for tracers
- units :
- meter2 second-1
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Akv_bak()float641e-05
- long_name :
- background vertical mixing coefficient for momentum
- units :
- meter2 second-1
array(1.e-05)
- rdrg()float640.000265
- long_name :
- linear drag coefficient
- units :
- meter second-1
array(0.000265)
- rdrg2()float640.003
- long_name :
- quadratic drag coefficient
array(0.003)
- Zob()float640.02
- long_name :
- bottom roughness
- units :
- meter
array(0.02)
- Zos()float640.02
- long_name :
- surface roughness
- units :
- meter
array(0.02)
- Znudg()float640.0
- long_name :
- free-surface nudging/relaxation inverse time scale
- units :
- day-1
array(0.)
- M2nudg()float640.0
- long_name :
- 2D momentum nudging/relaxation inverse time scale
- units :
- day-1
array(0.)
- M3nudg()float641.0
- long_name :
- 3D momentum nudging/relaxation inverse time scale
- units :
- day-1
array(1.)
- Tnudg(tracer)float64dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- Tracers nudging/relaxation inverse time scale
- units :
- day-1
Array Chunk Bytes 16 B 16 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - FSobc_in(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- free-surface inflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - FSobc_out(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- free-surface outflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M2obc_in(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 2D momentum inflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M2obc_out(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 2D momentum outflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Tobc_in(boundary, tracer)float64dask.array<chunksize=(4, 2), meta=np.ndarray>
- long_name :
- tracers inflow, nudging inverse time scale
- units :
- second-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 64 B 64 B Shape (4, 2) (4, 2) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Tobc_out(boundary, tracer)float64dask.array<chunksize=(4, 2), meta=np.ndarray>
- long_name :
- tracers outflow, nudging inverse time scale
- units :
- second-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 64 B 64 B Shape (4, 2) (4, 2) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M3obc_in(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 3D momentum inflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - M3obc_out(boundary)float64dask.array<chunksize=(4,), meta=np.ndarray>
- long_name :
- 3D momentum outflow, nudging inverse time scale
- units :
- second-1
Array Chunk Bytes 32 B 32 B Shape (4,) (4,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - rho0()float641.025e+03
- long_name :
- mean density used in Boussinesq approximation
- units :
- kilogram meter-3
array(1025.)
- gamma2()float641.0
- long_name :
- slipperiness parameter
array(1.)
- LuvSrc()int320
- long_name :
- momentum point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LwSrc()int320
- long_name :
- mass point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerSrc(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - LsshCLM()int320
- long_name :
- sea surface height climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Lm2CLM()int320
- long_name :
- 2D momentum climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Lm3CLM()int320
- long_name :
- 3D momentum climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerCLM(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - LnudgeM2CLM()int320
- long_name :
- 2D momentum climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LnudgeM3CLM()int320
- long_name :
- 3D momentum climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LnudgeTCLM(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - Lm2PSH()int320
- long_name :
- 2D momentum push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Lm3PSH()int320
- long_name :
- 3D momentum push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LtracerPSH(tracer)int32dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- tracer push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
Array Chunk Bytes 8 B 8 B Shape (2,) (2,) Count 3 Tasks 1 Chunks Type int32 numpy.ndarray - spherical()int321
- long_name :
- grid type logical switch
- flag_values :
- [0 1]
- flag_meanings :
- Cartesian spherical
array(1, dtype=int32)
- xl()float648.163e+06
- long_name :
- domain length in the XI-direction
- units :
- meter
array(8163221.23397392)
- el()float642.674e+06
- long_name :
- domain length in the ETA-direction
- units :
- meter
array(2674366.462091)
- Vtransform()int321
- long_name :
- vertical terrain-following transformation equation
array(1, dtype=int32)
- Vstretching()int321
- long_name :
- vertical terrain-following stretching function
array(1, dtype=int32)
- Tcline()float6475.0
- long_name :
- S-coordinate surface/bottom layer width
- units :
- meter
array(75.)
- hc()float6475.0
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
array(75.)
- grid()int321
- cf_role :
- grid_topology
- topology_dimension :
- 2
- node_dimensions :
- xi_psi eta_psi
- face_dimensions :
- xi_rho: xi_psi (padding: both) eta_rho: eta_psi (padding: both)
- edge1_dimensions :
- xi_u: xi_psi eta_u: eta_psi (padding: both)
- edge2_dimensions :
- xi_v: xi_psi (padding: both) eta_v: eta_psi
- node_coordinates :
- lon_psi lat_psi
- face_coordinates :
- lon_rho lat_rho
- edge1_coordinates :
- lon_u lat_u
- edge2_coordinates :
- lon_v lat_v
- vertical_dimensions :
- s_rho: s_w (padding: none)
array(1, dtype=int32)
- s_rho(s_rho)float64-0.99 -0.97 -0.95 ... -0.03 -0.01
- long_name :
- S-coordinate at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- standard_name :
- ocean_s_coordinate_g1
- formula_terms :
- s: s_rho C: Cs_r eta: zeta depth: h depth_c: hc
- field :
- s_rho, scalar
- axis :
- Z
array([-0.99, -0.97, -0.95, -0.93, -0.91, -0.89, -0.87, -0.85, -0.83, -0.81, -0.79, -0.77, -0.75, -0.73, -0.71, -0.69, -0.67, -0.65, -0.63, -0.61, -0.59, -0.57, -0.55, -0.53, -0.51, -0.49, -0.47, -0.45, -0.43, -0.41, -0.39, -0.37, -0.35, -0.33, -0.31, -0.29, -0.27, -0.25, -0.23, -0.21, -0.19, -0.17, -0.15, -0.13, -0.11, -0.09, -0.07, -0.05, -0.03, -0.01]) - s_w(s_w)float64-1.0 -0.98 -0.96 ... -0.02 0.0
- long_name :
- S-coordinate at W-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- standard_name :
- ocean_s_coordinate_g1
- formula_terms :
- s: s_w C: Cs_w eta: zeta depth: h depth_c: hc
- field :
- s_w, scalar
- axis :
- Z
array([-1. , -0.98, -0.96, -0.94, -0.92, -0.9 , -0.88, -0.86, -0.84, -0.82, -0.8 , -0.78, -0.76, -0.74, -0.72, -0.7 , -0.68, -0.66, -0.64, -0.62, -0.6 , -0.58, -0.56, -0.54, -0.52, -0.5 , -0.48, -0.46, -0.44, -0.42, -0.4 , -0.38, -0.36, -0.34, -0.32, -0.3 , -0.28, -0.26, -0.24, -0.22, -0.2 , -0.18, -0.16, -0.14, -0.12, -0.1 , -0.08, -0.06, -0.04, -0.02, 0. ]) - Cs_r(s_rho)float64dask.array<chunksize=(50,), meta=np.ndarray>
- long_name :
- S-coordinate stretching curves at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- field :
- Cs_r, scalar
Array Chunk Bytes 400 B 400 B Shape (50,) (50,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - Cs_w(s_w)float64dask.array<chunksize=(51,), meta=np.ndarray>
- long_name :
- S-coordinate stretching curves at W-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- field :
- Cs_w, scalar
Array Chunk Bytes 408 B 408 B Shape (51,) (51,) Count 3 Tasks 1 Chunks Type float64 numpy.ndarray - h(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- bathymetry at RHO-points
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- bath, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - f(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- Coriolis parameter at RHO-points
- units :
- second-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- coriolis, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - pm(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in XI
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pm, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - pn(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in ETA
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pn, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - lon_rho(lon_rho)float64-170.0 -170.0 ... -95.05 -95.0
- long_name :
- longitude of RHO-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_rho, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- X
array([-170. , -169.95, -169.9 , ..., -95.1 , -95.05, -95. ])
- lat_rho(lat_rho)float64-12.0 -11.95 -11.9 ... 11.95 12.0
- long_name :
- latitude of RHO-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_rho, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- Y
array([-11.999981, -11.949983, -11.899983, ..., 11.899983, 11.949983, 11.999981]) - lon_u(lon_u)float64-170.0 -169.9 ... -95.07 -95.02
- long_name :
- longitude of U-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_u, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- X
array([-169.975, -169.925, -169.875, ..., -95.125, -95.075, -95.025])
- lat_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- latitude of U-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_u, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 76.96 kB Shape (481, 1500) (481, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - lon_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- longitude of V-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_v, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - lat_v(lat_v)float64-11.97 -11.92 ... 11.92 11.97
- long_name :
- latitude of V-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_v, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- axis :
- Y
array([-11.974982, -11.924983, -11.874983, ..., 11.874983, 11.924983, 11.974982]) - lon_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- longitude of PSI-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_psi, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1500) (480, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - lat_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- latitude of PSI-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_psi, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1500) (480, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - angle(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- angle between XI-axis and EAST
- units :
- radians
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- angle, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - mask_rho(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- mask on RHO-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - mask_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- mask on U-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
Array Chunk Bytes 5.77 MB 76.96 kB Shape (481, 1500) (481, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - mask_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- mask on V-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 77 Tasks 76 Chunks Type float64 numpy.ndarray - mask_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- long_name :
- mask on psi-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- node
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1500) (480, 20) Count 76 Tasks 75 Chunks Type float64 numpy.ndarray - ocean_time(ocean_time)datetime64[ns]1985-10-02T03:00:00 ... 1985-11-...
- axis :
- T
- standard_name :
- time
array(['1985-10-02T03:00:00.000000000', '1985-10-02T09:00:00.000000000', '1985-10-02T15:00:00.000000000', '1985-10-02T21:00:00.000000000', '1985-10-03T03:00:00.000000000', '1985-10-03T09:00:00.000000000', '1985-10-03T15:00:00.000000000', '1985-10-03T21:00:00.000000000', '1985-10-04T03:00:00.000000000', '1985-10-04T09:00:00.000000000', '1985-10-04T15:00:00.000000000', '1985-10-04T21:00:00.000000000', '1985-10-05T03:00:00.000000000', '1985-10-05T09:00:00.000000000', '1985-10-05T15:00:00.000000000', '1985-10-05T21:00:00.000000000', '1985-10-06T03:00:00.000000000', '1985-10-06T09:00:00.000000000', '1985-10-06T15:00:00.000000000', '1985-10-06T21:00:00.000000000', '1985-10-07T03:00:00.000000000', '1985-10-07T09:00:00.000000000', '1985-10-07T15:00:00.000000000', '1985-10-07T21:00:00.000000000', '1985-10-08T03:00:00.000000000', '1985-10-08T09:00:00.000000000', '1985-10-08T15:00:00.000000000', '1985-10-08T21:00:00.000000000', '1985-10-09T03:00:00.000000000', '1985-10-09T09:00:00.000000000', '1985-10-09T15:00:00.000000000', '1985-10-09T21:00:00.000000000', '1985-10-10T03:00:00.000000000', '1985-10-10T09:00:00.000000000', '1985-10-10T15:00:00.000000000', '1985-10-10T21:00:00.000000000', '1985-10-11T03:00:00.000000000', '1985-10-11T09:00:00.000000000', '1985-10-11T15:00:00.000000000', '1985-10-11T21:00:00.000000000', '1985-10-12T03:00:00.000000000', '1985-10-12T09:00:00.000000000', '1985-10-12T15:00:00.000000000', '1985-10-12T21:00:00.000000000', '1985-10-13T03:00:00.000000000', '1985-10-13T09:00:00.000000000', '1985-10-13T15:00:00.000000000', '1985-10-13T21:00:00.000000000', '1985-10-14T03:00:00.000000000', '1985-10-14T09:00:00.000000000', '1985-10-14T15:00:00.000000000', '1985-10-14T21:00:00.000000000', '1985-10-15T03:00:00.000000000', '1985-10-15T09:00:00.000000000', '1985-10-15T15:00:00.000000000', '1985-10-15T21:00:00.000000000', '1985-10-16T03:00:00.000000000', '1985-10-16T09:00:00.000000000', '1985-10-16T15:00:00.000000000', '1985-10-16T21:00:00.000000000', '1985-10-17T03:00:00.000000000', '1985-10-17T09:00:00.000000000', '1985-10-17T15:00:00.000000000', '1985-10-17T21:00:00.000000000', '1985-10-18T03:00:00.000000000', '1985-10-18T09:00:00.000000000', '1985-10-18T15:00:00.000000000', '1985-10-18T21:00:00.000000000', '1985-10-19T03:00:00.000000000', '1985-10-19T09:00:00.000000000', '1985-10-19T15:00:00.000000000', '1985-10-19T21:00:00.000000000', '1985-10-20T03:00:00.000000000', '1985-10-20T09:00:00.000000000', '1985-10-20T15:00:00.000000000', '1985-10-20T21:00:00.000000000', '1985-10-21T03:00:00.000000000', '1985-10-21T09:00:00.000000000', '1985-10-21T15:00:00.000000000', '1985-10-21T21:00:00.000000000', '1985-10-22T03:00:00.000000000', '1985-10-22T09:00:00.000000000', '1985-10-22T15:00:00.000000000', '1985-10-22T21:00:00.000000000', '1985-10-23T03:00:00.000000000', '1985-10-23T09:00:00.000000000', '1985-10-23T15:00:00.000000000', '1985-10-23T21:00:00.000000000', '1985-10-24T03:00:00.000000000', '1985-10-24T09:00:00.000000000', '1985-10-24T15:00:00.000000000', '1985-10-24T21:00:00.000000000', '1985-10-25T03:00:00.000000000', '1985-10-25T09:00:00.000000000', '1985-10-25T15:00:00.000000000', '1985-10-25T21:00:00.000000000', '1985-10-26T03:00:00.000000000', '1985-10-26T09:00:00.000000000', '1985-10-26T15:00:00.000000000', '1985-10-26T21:00:00.000000000', '1985-10-27T03:00:00.000000000', '1985-10-27T09:00:00.000000000', '1985-10-27T15:00:00.000000000', '1985-10-27T21:00:00.000000000', '1985-10-28T03:00:00.000000000', '1985-10-28T09:00:00.000000000', '1985-10-28T15:00:00.000000000', '1985-10-28T21:00:00.000000000', '1985-10-29T03:00:00.000000000', '1985-10-29T09:00:00.000000000', '1985-10-29T15:00:00.000000000', '1985-10-29T21:00:00.000000000', '1985-10-30T03:00:00.000000000', '1985-10-30T09:00:00.000000000', '1985-10-30T15:00:00.000000000', '1985-10-30T21:00:00.000000000', '1985-10-31T03:00:00.000000000', '1985-10-31T09:00:00.000000000', '1985-10-31T15:00:00.000000000', '1985-10-31T21:00:00.000000000', '1985-11-01T03:00:00.000000000', '1985-11-01T09:00:00.000000000', '1985-11-01T15:00:00.000000000', '1985-11-01T21:00:00.000000000', '1985-11-02T03:00:00.000000000', '1985-11-02T09:00:00.000000000', '1985-11-02T15:00:00.000000000', '1985-11-02T21:00:00.000000000', '1985-11-03T03:00:00.000000000', '1985-11-03T09:00:00.000000000', '1985-11-03T15:00:00.000000000', '1985-11-03T21:00:00.000000000', '1985-11-04T03:00:00.000000000', '1985-11-04T09:00:00.000000000', '1985-11-04T15:00:00.000000000', '1985-11-04T21:00:00.000000000', '1985-11-05T03:00:00.000000000', '1985-11-05T09:00:00.000000000', '1985-11-05T15:00:00.000000000', '1985-11-05T21:00:00.000000000'], dtype='datetime64[ns]') - z_w(ocean_time, s_w, lat_rho, lon_rho)float64dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.24 GB 196.25 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 2368 Tasks 228 Chunks Type float64 numpy.ndarray - z_w_u(ocean_time, s_w, lat_rho, lon_u)float64dask.array<chunksize=(50, 51, 481, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.21 GB 186.44 MB Shape (140, 51, 481, 1500) (50, 51, 481, 19) Count 5515 Tasks 450 Chunks Type float64 numpy.ndarray - z_w_v(ocean_time, s_w, lat_v, lon_rho)float64dask.array<chunksize=(50, 51, 480, 20), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.15 GB 195.84 MB Shape (140, 51, 480, 1501) (50, 51, 480, 20) Count 3280 Tasks 228 Chunks Type float64 numpy.ndarray - z_w_psi(ocean_time, s_w, lat_v, lon_u)float64dask.array<chunksize=(50, 51, 480, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.13 GB 186.05 MB Shape (140, 51, 480, 1500) (50, 51, 480, 19) Count 7315 Tasks 450 Chunks Type float64 numpy.ndarray - z_rho(ocean_time, s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.43 GB 192.40 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 2368 Tasks 228 Chunks Type float64 numpy.ndarray - z_rho_u(ocean_time, s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.40 GB 182.78 MB Shape (140, 50, 481, 1500) (50, 50, 481, 19) Count 5515 Tasks 450 Chunks Type float64 numpy.ndarray - z_rho_v(ocean_time, s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.35 GB 192.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 3280 Tasks 228 Chunks Type float64 numpy.ndarray - z_rho_psi(ocean_time, s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.32 GB 182.40 MB Shape (140, 50, 480, 1500) (50, 50, 480, 19) Count 7315 Tasks 450 Chunks Type float64 numpy.ndarray - z_rho0(s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.79 MB 3.85 MB Shape (50, 481, 1501) (50, 481, 20) Count 315 Tasks 76 Chunks Type float64 numpy.ndarray - z_rho_u0(s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.60 MB 3.66 MB Shape (50, 481, 1500) (50, 481, 19) Count 1364 Tasks 150 Chunks Type float64 numpy.ndarray - z_rho_v0(s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.19 MB 3.84 MB Shape (50, 480, 1501) (50, 480, 20) Count 619 Tasks 76 Chunks Type float64 numpy.ndarray - z_rho_psi0(s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.00 MB 3.65 MB Shape (50, 480, 1500) (50, 480, 19) Count 1964 Tasks 150 Chunks Type float64 numpy.ndarray - z_w0(s_w, lat_rho, lon_rho)float64dask.array<chunksize=(51, 481, 20), meta=np.ndarray>
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.57 MB 3.92 MB Shape (51, 481, 1501) (51, 481, 20) Count 315 Tasks 76 Chunks Type float64 numpy.ndarray - z_w_u0(s_w, lat_rho, lon_u)float64dask.array<chunksize=(51, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.37 MB 3.73 MB Shape (51, 481, 1500) (51, 481, 19) Count 1364 Tasks 150 Chunks Type float64 numpy.ndarray - z_w_v0(s_w, lat_v, lon_rho)float64dask.array<chunksize=(51, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.96 MB 3.92 MB Shape (51, 480, 1501) (51, 480, 20) Count 619 Tasks 76 Chunks Type float64 numpy.ndarray - z_w_psi0(s_w, lat_v, lon_u)float64dask.array<chunksize=(51, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.76 MB 3.72 MB Shape (51, 480, 1500) (51, 480, 19) Count 1964 Tasks 150 Chunks Type float64 numpy.ndarray
- zeta(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged free-surface
- units :
- meter
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- free-surface, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - ubar(ocean_time, lat_rho, lon_u)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged vertically integrated u-momentum component
- units :
- meter second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- ubar-velocity, scalar, series
Array Chunk Bytes 404.04 MB 1.92 MB Shape (140, 481, 1500) (50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - vbar(ocean_time, lat_v, lon_rho)float32dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- long_name :
- time-averaged vertically integrated v-momentum component
- units :
- meter second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- vbar-velocity, scalar, series
Array Chunk Bytes 403.47 MB 1.92 MB Shape (140, 480, 1501) (50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 1), meta=np.ndarray>
Array Chunk Bytes 20.22 GB 91.39 MB Shape (140, 50, 481, 1501) (50, 50, 481, 19) Count 3400 Tasks 453 Chunks Type float32 numpy.ndarray - v(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 1, 20), meta=np.ndarray>
Array Chunk Bytes 20.22 GB 95.80 MB Shape (140, 50, 481, 1501) (50, 50, 479, 20) Count 5245 Tasks 684 Chunks Type float32 numpy.ndarray - w(ocean_time, s_w, lat_rho, lon_rho)float32dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged vertical momentum component
- units :
- meter second-1
- time :
- ocean_time
- standard_name :
- upward_sea_water_velocity
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- w-velocity, scalar, series
Array Chunk Bytes 20.62 GB 98.12 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged potential temperature
- units :
- Celsius
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temperature, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged salinity
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salinity, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - Hsbl(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged depth of oceanic surface boundary layer
- units :
- meter
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- Hsbl, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - AKv(ocean_time, s_w, lat_rho, lon_rho)float32dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged vertical viscosity coefficient
- units :
- meter2 second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- AKv, scalar, series
Array Chunk Bytes 20.62 GB 98.12 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - AKt(ocean_time, s_w, lat_rho, lon_rho)float32dask.array<chunksize=(50, 51, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged temperature vertical diffusion coefficient
- units :
- meter2 second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- AKt, scalar, series
Array Chunk Bytes 20.62 GB 98.12 MB Shape (140, 51, 481, 1501) (50, 51, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - shflux(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged surface net heat flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- surface heat flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - ssflux(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged surface net salt flux, (E-P)*SALT
- units :
- meter second-1
- negative_value :
- upward flux, freshening (net precipitation)
- positive_value :
- downward flux, salting (net evaporation)
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- surface net salt flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - latent(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged net latent heat flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- latent heat flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - sensible(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged net sensible heat flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- sensible heat flux, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - lwrad(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged net longwave radiation flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- longwave radiation, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - evaporation(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged evaporation rate
- units :
- kilogram meter-2 second-1
- negative_value :
- downward flux, freshening (condensation)
- positive_value :
- upward flux, salting (evaporation)
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- evaporation, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - rain(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged rain fall rate
- units :
- kilogram meter-2 second-1
- positive_value :
- downward flux, freshening (precipitation)
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- rain, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - swrad(ocean_time, lat_rho, lon_rho)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged solar shortwave radiation flux
- units :
- watt meter-2
- negative_value :
- upward flux, cooling
- positive_value :
- downward flux, heating
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- shortwave radiation, scalar, series
Array Chunk Bytes 404.31 MB 1.92 MB Shape (140, 481, 1501) (50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - sustr(ocean_time, lat_rho, lon_u)float32dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- long_name :
- time-averaged surface u-momentum stress
- units :
- newton meter-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- surface u-momentum stress, scalar, series
Array Chunk Bytes 404.04 MB 1.92 MB Shape (140, 481, 1500) (50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - svstr(ocean_time, lat_v, lon_rho)float32dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- long_name :
- time-averaged surface v-momentum stress
- units :
- newton meter-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- surface v-momentum stress, scalar, series
Array Chunk Bytes 403.47 MB 1.92 MB Shape (140, 480, 1501) (50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_cor(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, Coriolis term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_cor, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_cor(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, Coriolis term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_cor, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_vadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, vertical advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_vadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_vadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, vertical advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_vadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_hadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_hadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_hadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_hadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_xadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal XI-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_xadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_xadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal XI-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_xadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_yadv(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal ETA-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_yadv, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_yadv(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal ETA-advection term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_yadv, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_prsgrd(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, pressure gradient term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_prsgrd, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_prsgrd(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, pressure gradient term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_prsgrd, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_vvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, vertical viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_vvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_vvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, vertical viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_vvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_hvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_hvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_hvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_hvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_xvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal XI-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_xvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_xvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal XI-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_xvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_yvisc(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, horizontal ETA-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_yvisc, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_yvisc(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, horizontal ETA-viscosity term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_yvisc, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - u_accel(ocean_time, s_rho, lat_rho, lon_u)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- 3D u-momentum, acceleration term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
- field :
- u_accel, scalar, series
Array Chunk Bytes 20.20 GB 96.20 MB Shape (140, 50, 481, 1500) (50, 50, 481, 20) Count 226 Tasks 225 Chunks Type float32 numpy.ndarray - v_accel(ocean_time, s_rho, lat_v, lon_rho)float32dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- long_name :
- 3D v-momentum, acceleration term
- units :
- meter second-2
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
- field :
- v_accel, scalar, series
Array Chunk Bytes 20.17 GB 96.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_hadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_hadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_xadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal XI-advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_xadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_yadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal ETA-advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_yadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_vadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, vertical advection term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_vadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_hdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_hdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_xdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal XI-diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_xdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_ydiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal ETA-diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_ydiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_sdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, horizontal S-diffusion rotated tensor term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_sdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_vdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, vertical diffusion term
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_vdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - temp_rate(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- potential temperature, time rate of change
- units :
- Celsius second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- temp_rate, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_hadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_hadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_xadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal XI-advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_xadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_yadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal ETA-advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_yadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_vadv(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, vertical advection term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_vadv, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_hdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_hdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_xdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal XI-diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_xdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_ydiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal ETA-diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_ydiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_sdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, horizontal S-diffusion rotated tensor term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_sdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_vdiff(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, vertical diffusion term
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_vdiff, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - salt_rate(ocean_time, s_rho, lat_rho, lon_rho)float32dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- long_name :
- salinity, time rate of change
- units :
- nondimensional second-1
- time :
- ocean_time
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- salt_rate, scalar, series
Array Chunk Bytes 20.22 GB 96.20 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 229 Tasks 228 Chunks Type float32 numpy.ndarray - pm_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 381 Tasks 76 Chunks Type float64 numpy.ndarray - pn_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1126 Tasks 150 Chunks Type float64 numpy.ndarray - pm_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1126 Tasks 150 Chunks Type float64 numpy.ndarray - pn_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 381 Tasks 76 Chunks Type float64 numpy.ndarray - pm_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1430 Tasks 150 Chunks Type float64 numpy.ndarray - pn_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1726 Tasks 150 Chunks Type float64 numpy.ndarray - dx(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in XI
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pm, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 153 Tasks 76 Chunks Type float64 numpy.ndarray - dx_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1276 Tasks 150 Chunks Type float64 numpy.ndarray - dx_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 457 Tasks 76 Chunks Type float64 numpy.ndarray - dx_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1580 Tasks 150 Chunks Type float64 numpy.ndarray - dy(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in ETA
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pn, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 153 Tasks 76 Chunks Type float64 numpy.ndarray - dy_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 1276 Tasks 150 Chunks Type float64 numpy.ndarray - dy_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 457 Tasks 76 Chunks Type float64 numpy.ndarray - dy_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 1876 Tasks 150 Chunks Type float64 numpy.ndarray - dz(ocean_time, s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.43 GB 192.40 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 3052 Tasks 228 Chunks Type float64 numpy.ndarray - dz_w(ocean_time, s_w, lat_rho, lon_rho)float64dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.24 GB 188.55 MB Shape (140, 51, 481, 1501) (50, 49, 481, 20) Count 6472 Tasks 684 Chunks Type float64 numpy.ndarray - dz_u(ocean_time, s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.40 GB 182.78 MB Shape (140, 50, 481, 1500) (50, 50, 481, 19) Count 6199 Tasks 450 Chunks Type float64 numpy.ndarray - dz_w_u(ocean_time, s_w, lat_rho, lon_u)float64dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.21 GB 179.12 MB Shape (140, 51, 481, 1500) (50, 49, 481, 19) Count 15913 Tasks 1350 Chunks Type float64 numpy.ndarray - dz_v(ocean_time, s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.35 GB 192.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 3964 Tasks 228 Chunks Type float64 numpy.ndarray - dz_w_v(ocean_time, s_w, lat_v, lon_rho)float64dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.15 GB 188.16 MB Shape (140, 51, 480, 1501) (50, 49, 480, 20) Count 9208 Tasks 684 Chunks Type float64 numpy.ndarray - dz_psi(ocean_time, s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.32 GB 182.40 MB Shape (140, 50, 480, 1500) (50, 50, 480, 19) Count 7111 Tasks 450 Chunks Type float64 numpy.ndarray - dz_w_psi(ocean_time, s_w, lat_v, lon_u)float64dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.13 GB 178.75 MB Shape (140, 51, 480, 1500) (50, 49, 480, 19) Count 18649 Tasks 1350 Chunks Type float64 numpy.ndarray - dz0(s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.79 MB 3.85 MB Shape (50, 481, 1501) (50, 481, 20) Count 543 Tasks 76 Chunks Type float64 numpy.ndarray - dz_w0(s_w, lat_rho, lon_rho)float64dask.array<chunksize=(1, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.57 MB 3.77 MB Shape (51, 481, 1501) (49, 481, 20) Count 1683 Tasks 228 Chunks Type float64 numpy.ndarray - dz_u0(s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.60 MB 3.66 MB Shape (50, 481, 1500) (50, 481, 19) Count 1592 Tasks 150 Chunks Type float64 numpy.ndarray - dz_w_u0(s_w, lat_rho, lon_u)float64dask.array<chunksize=(1, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 294.37 MB 3.58 MB Shape (51, 481, 1500) (49, 481, 19) Count 4830 Tasks 450 Chunks Type float64 numpy.ndarray - dz_v0(s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.19 MB 3.84 MB Shape (50, 480, 1501) (50, 480, 20) Count 847 Tasks 76 Chunks Type float64 numpy.ndarray - dz_w_v0(s_w, lat_v, lon_rho)float64dask.array<chunksize=(1, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.96 MB 3.76 MB Shape (51, 480, 1501) (49, 480, 20) Count 2595 Tasks 228 Chunks Type float64 numpy.ndarray - dz_psi0(s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 288.00 MB 3.65 MB Shape (50, 480, 1500) (50, 480, 19) Count 1896 Tasks 150 Chunks Type float64 numpy.ndarray - dz_w_psi0(s_w, lat_v, lon_u)float64dask.array<chunksize=(1, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 293.76 MB 3.58 MB Shape (51, 480, 1500) (49, 480, 19) Count 5742 Tasks 450 Chunks Type float64 numpy.ndarray - dA(lat_rho, lon_rho)float64dask.array<chunksize=(481, 20), meta=np.ndarray>
- long_name :
- curvilinear coordinate metric in XI
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pm, scalar
Array Chunk Bytes 5.78 MB 76.96 kB Shape (481, 1501) (481, 20) Count 382 Tasks 76 Chunks Type float64 numpy.ndarray - dA_u(lat_rho, lon_u)float64dask.array<chunksize=(481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.77 MB 73.11 kB Shape (481, 1500) (481, 19) Count 2702 Tasks 150 Chunks Type float64 numpy.ndarray - dA_v(lat_v, lon_rho)float64dask.array<chunksize=(480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 76.80 kB Shape (480, 1501) (480, 20) Count 990 Tasks 76 Chunks Type float64 numpy.ndarray - dA_psi(lat_v, lon_u)float64dask.array<chunksize=(480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 5.76 MB 72.96 kB Shape (480, 1500) (480, 19) Count 3606 Tasks 150 Chunks Type float64 numpy.ndarray - dV(ocean_time, s_rho, lat_rho, lon_rho)float64dask.array<chunksize=(50, 50, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.43 GB 192.40 MB Shape (140, 50, 481, 1501) (50, 50, 481, 20) Count 3966 Tasks 228 Chunks Type float64 numpy.ndarray - dV_w(ocean_time, s_w, lat_rho, lon_rho)float64dask.array<chunksize=(50, 1, 481, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.24 GB 188.55 MB Shape (140, 51, 481, 1501) (50, 49, 481, 20) Count 8298 Tasks 684 Chunks Type float64 numpy.ndarray - dV_u(ocean_time, s_rho, lat_rho, lon_u)float64dask.array<chunksize=(50, 50, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.40 GB 182.78 MB Shape (140, 50, 481, 1500) (50, 50, 481, 19) Count 9951 Tasks 450 Chunks Type float64 numpy.ndarray - dV_w_u(ocean_time, s_w, lat_rho, lon_u)float64dask.array<chunksize=(50, 1, 481, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.21 GB 179.12 MB Shape (140, 51, 481, 1500) (50, 49, 481, 19) Count 21465 Tasks 1350 Chunks Type float64 numpy.ndarray - dV_v(ocean_time, s_rho, lat_v, lon_rho)float64dask.array<chunksize=(50, 50, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.35 GB 192.00 MB Shape (140, 50, 480, 1501) (50, 50, 480, 20) Count 5486 Tasks 228 Chunks Type float64 numpy.ndarray - dV_w_v(ocean_time, s_w, lat_v, lon_rho)float64dask.array<chunksize=(50, 1, 480, 20), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.15 GB 188.16 MB Shape (140, 51, 480, 1501) (50, 49, 480, 20) Count 11642 Tasks 684 Chunks Type float64 numpy.ndarray - dV_psi(ocean_time, s_rho, lat_v, lon_u)float64dask.array<chunksize=(50, 50, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 40.32 GB 182.40 MB Shape (140, 50, 480, 1500) (50, 50, 480, 19) Count 11767 Tasks 450 Chunks Type float64 numpy.ndarray - dV_w_psi(ocean_time, s_w, lat_v, lon_u)float64dask.array<chunksize=(50, 1, 480, 19), meta=np.ndarray>
- grid :
- <xgcm.Grid> X Axis (not periodic, boundary=None): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic, boundary=None): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic, boundary=None): * center s_rho --> outer * outer s_w --> center
Array Chunk Bytes 41.13 GB 178.75 MB Shape (140, 51, 480, 1500) (50, 49, 480, 19) Count 25105 Tasks 1350 Chunks Type float64 numpy.ndarray
- file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- format :
- netCDF-3 64bit offset file
- Conventions :
- CF-1.4, SGRID-0.3
- type :
- ROMS/TOMS diagnostics file
- title :
- TPOS 1/20deg
- var_info :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/varinfo.dat
- rst_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2_2.nc
- avg_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2_2.nc
- dia_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- sta_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2_2.nc
- grd_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_grd.nc
- ini_file :
- /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
- frc_file_01 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_ncep.nc
- frc_file_02 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_giss.nc
- frc_file_03 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_gcgcs.nc
- bry_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_bry.nc
- script_file :
- ./tpos20_3_diag_2.in
- spos_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/stations_tpos20.in
- NLM_LBC :
- EDGE: WEST SOUTH EAST NORTH zeta: Che Che Che Che ubar: Shc Shc Shc Shc vbar: Shc Shc Shc Shc u: RadNud RadNud RadNud RadNud v: RadNud RadNud RadNud RadNud temp: RadNud RadNud Cla RadNud salt: RadNud RadNud Cla RadNud
- svn_url :
- https:://myroms.org/svn/src
- svn_rev :
- Unversioned directory
- code_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROMS/ROMS
- header_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INC/
- header_file :
- tpos20.h
- os :
- Linux
- cpu :
- x86_64
- compiler_system :
- ifort
- compiler_command :
- /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/mpi/mpif90
- compiler_flags :
- -fp-model precise -ip -O3
- tiling :
- 018x012
- history :
- ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:53:43 PM
- ana_file :
- ROMS/Functionals/ana_btflux.h
- CPP_options :
- TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SHAPE, AVERAGES, !BOUNDARY_A BULK_FLUXES, CCSM_FLUXES, !COLLECT_ALL..., CURVGRID, DIAGNOSTICS_TS, DIAGNOSTICS_UV, DJ_GRADPS, DOUBLE_PRECISION, EMINUSP, INLINE_2DIO, LMD_CONVEC, LMD_DDMIX, LMD_MIXING, LMD_NONLOCAL, LMD_RIMIX, LMD_SHAPIRO, LMD_SKPP, LONGWAVE_OUT, MASKING, MIX_ISO_TS, MIX_S_UV, MPI, NONLINEAR, NONLIN_EOS, NO_LBC_ATT, PERFECT_RESTART, POWER_LAW, PROFILE, RADIATION_2D, REDUCE_ALLGATHER, RI_SPLINES, !RST_SINGLE, SALINITY, SOLAR_SOURCE, SOLVE3D, SPLINES_VDIFF, SPLINES_VVISC, STATIONS, TS_U3HADVECTION, TS_C4VADVECTION, TS_DIF4, UV_ADV, UV_COR, UV_U3HADVECTION, UV_C4VADVECTION, UV_QDRAG, UV_VIS4, VAR_RHO_2D
t0 = subsetter(ds).ocean_time[1].data # pd.Timestamp("1985-Oct-02 00:00:00").to_numpy()
subds = ds.pipe(subsetter).pipe(interpolate, newz).rename(renamer)
RHS forcing#
rhs = xr.Dataset()
rhs_tracer = ["hadv", "vadv", "hdiff"]
rhs_mom = ["prsgrd", "hadv", "vadv", "hvisc"]
for var, terms in zip(
["u", "v", "temp", "salt"], [rhs_mom, rhs_mom, rhs_tracer, rhs_tracer]
):
varnames = [f"{var}_{term}" for term in terms]
rhs[var] = (subds[varnames].to_array("term").sum("term")) * 86400
rhs[var].attrs["description"] = f"sum over {varnames}"
rhs
<xarray.Dataset>
Dimensions: (ocean_time: 20, z_rho: 216)
Coordinates:
nDIA int32 180
LwSrc int32 0
Zos float64 0.02
Fgamma float64 0.284
lon_u float64 -140.0
Zob float64 0.02
ntsDIA int32 1
rdrg float64 0.000265
mask_rho float64 1.0
z_rho_psi (ocean_time, z_rho) float64 -107.8 -107.3 ... -0.7773 -0.2768
gamma2 float64 1.0
f float64 4.514e-18
ndefAVG int32 0
dt float64 120.0
z_rho_v (ocean_time, z_rho) float64 -107.7 -107.2 ... -0.776 -0.2764
ntimes int32 25201
lon_rho float64 -140.0
ndtfast int32 20
mask_u float64 1.0
lon_v float64 -140.0
Falpha float64 2.0
M2nudg float64 0.0
* z_rho (z_rho) float64 -107.8 -107.2 -106.8 ... -1.25 -0.75 -0.25
LuvSrc int32 0
dstart object 0027-09-02 00:00:00
ntsAVG int32 1
xl float64 8.163e+06
Vtransform int32 1
dtfast float64 6.0
Lm2PSH int32 0
pn float64 0.0001799
LsshCLM int32 0
LnudgeM2CLM int32 0
mask_v float64 1.0
rdrg2 float64 0.003
z_rho0 (z_rho) float64 -108.1 -107.6 -107.1 ... -1.577 -1.077 -0.5769
lat_u float64 1.772e-12
LnudgeM3CLM int32 0
Cs_r (z_rho) float64 -0.02114 -0.02104 ... -0.0002008 -0.0001072
rho0 float64 1.025e+03
hc float64 75.0
nHIS int32 3600
ndefDIA int32 0
el float64 2.674e+06
angle float64 4.106e-14
pm float64 0.0001799
Znudg float64 0.0
spherical int32 1
ndefHIS int32 0
Akv_bak float64 1e-05
z_rho_v0 (z_rho) float64 -108.0 -107.5 -107.0 ... -1.576 -1.076 -0.5765
nl_visc4 float64 1.24e+09
grid int32 1
z_rho_u0 (z_rho) float64 -108.2 -107.7 -107.2 ... -1.578 -1.078 -0.5774
nSTA int32 30
M3nudg float64 1.0
nRST int32 3600
Tcline float64 75.0
Lm2CLM int32 0
Lm3PSH int32 0
mask_psi float64 1.0
Lm3CLM int32 0
lat_psi float64 -0.025
s_rho (z_rho) float64 -0.2463 -0.2455 -0.2446 ... -0.003013 -0.001633
z_rho_u (ocean_time, z_rho) float64 -107.8 -107.3 ... -0.777 -0.2766
* ocean_time (ocean_time) datetime64[ns] 1985-10-02T03:00:00 ... 1985-10-...
lat_rho float64 1.45e-14
LuvSponge int32 0
nAVG int32 180
lat_v float64 -0.025
h float64 4.314e+03
z_rho_psi0 (z_rho) float64 -108.2 -107.7 -107.2 ... -1.578 -1.078 -0.5774
lon_psi float64 -140.0
Vstretching int32 1
Data variables:
u (ocean_time, z_rho) float64 0.0935 0.09244 ... 0.1627 0.1625
v (ocean_time, z_rho) float64 0.106 0.1083 ... -0.0536 -0.05361
temp (ocean_time, z_rho) float64 -0.8086 -0.7765 ... -0.02899
salt (ocean_time, z_rho) float64 -0.02245 -0.02013 ... 0.0006571- ocean_time: 20
- z_rho: 216
- nDIA()int32180
- long_name :
- number of time-steps between diagnostic records
array(180, dtype=int32)
- LwSrc()int320
- long_name :
- mass point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Zos()float640.02
- long_name :
- surface roughness
- units :
- meter
array(0.02)
- Fgamma()float640.284
- long_name :
- Power-law shape barotropic filter parameter
array(0.284)
- lon_u()float64-140.0
- long_name :
- longitude of U-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_u, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- axis :
- X
array(-140.025)
- Zob()float640.02
- long_name :
- bottom roughness
- units :
- meter
array(0.02)
- ntsDIA()int321
- long_name :
- starting time-step for accumulation of diagnostic fields
array(1, dtype=int32)
- rdrg()float640.000265
- long_name :
- linear drag coefficient
- units :
- meter second-1
array(0.000265)
- mask_rho()float641.0
- long_name :
- mask on RHO-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- face
array(1.)
- z_rho_psi(ocean_time, z_rho)float64-107.8 -107.3 ... -0.7773 -0.2768
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array([[-107.84525484, -107.34477137, -106.8442879 , ..., -1.24743169, -0.74699274, -0.24655378], [-107.84175865, -107.34127477, -106.84079088, ..., -1.24384699, -0.74340761, -0.24296824], [-107.84104663, -107.34056266, -106.84007869, ..., -1.24311694, -0.74267748, -0.24223803], ..., [-107.86471398, -107.36423282, -106.86375167, ..., -1.26738349, -0.76694685, -0.2665102 ], [-107.86840759, -107.36792687, -106.86744616, ..., -1.27117061, -0.77073441, -0.2702982 ], [-107.87479259, -107.37431263, -106.87383267, ..., -1.27771727, -0.77728182, -0.27684637]]) - gamma2()float641.0
- long_name :
- slipperiness parameter
array(1.)
- f()float644.514e-18
- long_name :
- Coriolis parameter at RHO-points
- units :
- second-1
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- coriolis, scalar
array(4.51402673e-18)
- ndefAVG()int320
- long_name :
- number of time-steps between the creation of average files
array(0, dtype=int32)
- dt()float64120.0
- long_name :
- size of long time-steps
- units :
- second
array(120.)
- z_rho_v(ocean_time, z_rho)float64-107.7 -107.2 ... -0.776 -0.2764
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array([[-107.6703521 , -107.17072211, -106.67109213, ..., -1.24521184, -0.74554772, -0.2458836 ], [-107.66705257, -107.1674222 , -106.66779182, ..., -1.24182875, -0.74216424, -0.24249973], [-107.6659892 , -107.1663587 , -106.6667282 , ..., -1.24073845, -0.74107381, -0.24140917], ..., [-107.68976102, -107.19013335, -106.69050567, ..., -1.2651123 , -0.76545049, -0.26578868], [-107.69348754, -107.19386031, -106.69423308, ..., -1.2689332 , -0.76927183, -0.26961046], [-107.70007924, -107.20045279, -106.70082634, ..., -1.27569183, -0.77603125, -0.27637066]]) - ntimes()int3225201
- long_name :
- number of long time-steps
array(25201, dtype=int32)
- lon_rho()float64-140.0
- long_name :
- longitude of RHO-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_rho, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- axis :
- X
array(-140.)
- ndtfast()int3220
- long_name :
- number of short time-steps
array(20, dtype=int32)
- mask_u()float641.0
- long_name :
- mask on U-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- edge1
array(1.)
- lon_v()float64-140.0
- long_name :
- longitude of V-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_v, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array(-140.)
- Falpha()float642.0
- long_name :
- Power-law shape barotropic filter parameter
array(2.)
- M2nudg()float640.0
- long_name :
- 2D momentum nudging/relaxation inverse time scale
- units :
- day-1
array(0.)
- z_rho(z_rho)float64-107.8 -107.2 ... -0.75 -0.25
array([-107.75, -107.25, -106.75, ..., -1.25, -0.75, -0.25])
- LuvSrc()int320
- long_name :
- momentum point sources and sink activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- dstart()object0027-09-02 00:00:00
- long_name :
- time stamp assigned to model initilization
array(cftime.DatetimeNoLeap(27, 9, 2, 0, 0, 0, 0), dtype=object)
- ntsAVG()int321
- long_name :
- starting time-step for accumulation of time-averaged fields
array(1, dtype=int32)
- xl()float648.163e+06
- long_name :
- domain length in the XI-direction
- units :
- meter
array(8163221.23397392)
- Vtransform()int321
- long_name :
- vertical terrain-following transformation equation
array(1, dtype=int32)
- dtfast()float646.0
- long_name :
- size of short time-steps
- units :
- second
array(6.)
- Lm2PSH()int320
- long_name :
- 2D momentum push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- pn()float640.0001799
- long_name :
- curvilinear coordinate metric in ETA
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pn, scalar
array(0.00017986)
- LsshCLM()int320
- long_name :
- sea surface height climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- LnudgeM2CLM()int320
- long_name :
- 2D momentum climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- mask_v()float641.0
- long_name :
- mask on V-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- edge2
array(1.)
- rdrg2()float640.003
- long_name :
- quadratic drag coefficient
array(0.003)
- z_rho0(z_rho)float64-108.1 -107.6 ... -1.077 -0.5769
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array([-108.0687844 , -107.5688223 , -107.0688602 , -106.5688981 , -106.06893599, -105.56897389, -105.06901179, -104.56904969, -104.06908759, -103.56912548, -103.06916338, -102.56920128, -102.06923918, -101.56927708, -101.06931497, -100.56935287, -100.06939077, -99.56942867, -99.06946657, -98.56950446, -98.06954236, -97.56958026, -97.06961816, -96.56965606, -96.06969395, -95.56973185, -95.06976975, -94.56980765, -94.06984554, -93.56988344, -93.06992134, -92.56995924, -92.06999714, -91.57003503, -91.07007293, -90.57011083, -90.07014873, -89.57018663, -89.07022452, -88.57026242, -88.07030032, -87.57033822, -87.07037612, -86.57041401, -86.07045191, -85.57048981, -85.07052771, -84.57056561, -84.0706035 , -83.5706414 , -83.0706793 , -82.5707172 , -82.0707551 , -81.57079299, -81.07083089, -80.57086879, -80.07090669, -79.57094459, -79.07098248, -78.57102038, -78.07105828, -77.57109618, -77.07113408, -76.57117197, -76.07120987, -75.57124777, -75.07128567, -74.57132356, -74.07136146, -73.57139936, -73.07143726, -72.57147516, -72.07151305, -71.57155095, -71.07158885, -70.57162675, -70.07166465, -69.57170254, -69.07174044, -68.57177834, ... -40.07393852, -39.57397642, -39.07401432, -38.57405222, -38.07409011, -37.57412801, -37.07416591, -36.57420381, -36.07424171, -35.5742796 , -35.0743175 , -34.5743554 , -34.0743933 , -33.5744312 , -33.07446909, -32.57450699, -32.07454489, -31.57458279, -31.07462069, -30.57465858, -30.07469648, -29.57473438, -29.07477228, -28.57481018, -28.07484807, -27.57488597, -27.07492387, -26.57496177, -26.07499967, -25.57503756, -25.07507546, -24.57511336, -24.07515126, -23.57518916, -23.07522705, -22.57526495, -22.07530285, -21.57534075, -21.07537865, -20.57541654, -20.07545444, -19.57549234, -19.07553024, -18.57556813, -18.07560603, -17.57564393, -17.07568183, -16.57571973, -16.07575762, -15.57579552, -15.07583342, -14.57587132, -14.07590922, -13.57594711, -13.07598501, -12.57602291, -12.07606081, -11.57609871, -11.0761366 , -10.5761745 , -10.0762124 , -9.5762503 , -9.0762882 , -8.57632609, -8.07636399, -7.57640189, -7.07643979, -6.57647769, -6.07651558, -5.57655348, -5.07659138, -4.57662928, -4.07666718, -3.57670507, -3.07674297, -2.57678087, -2.07681877, -1.57685667, -1.07689456, -0.57693246]) - lat_u()float641.772e-12
- long_name :
- latitude of U-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_u, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array(1.77225986e-12)
- LnudgeM3CLM()int320
- long_name :
- 3D momentum climatology nudging activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Cs_r(z_rho)float64-0.02114 -0.02104 ... -0.0001072
- long_name :
- S-coordinate stretching curves at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- field :
- Cs_r, scalar
array([-0.02113809, -0.02103507, -0.02093205, -0.02082903, -0.02072601, -0.02062299, -0.02051997, -0.02041695, -0.02031392, -0.0202109 , -0.02010788, -0.02000486, -0.01990184, -0.01979882, -0.0196958 , -0.01959278, -0.01948976, -0.01938674, -0.01928372, -0.0191807 , -0.01907842, -0.0189765 , -0.01887458, -0.01877266, -0.01867074, -0.01856882, -0.0184669 , -0.01836497, -0.01826305, -0.01816113, -0.01805921, -0.01795729, -0.01785537, -0.01775345, -0.01765153, -0.01754961, -0.01744768, -0.01734576, -0.01724384, -0.01714192, -0.01704 , -0.01693808, -0.01683683, -0.01673602, -0.01663522, -0.01653441, -0.0164336 , -0.01633279, -0.01623199, -0.01613118, -0.01603037, -0.01592957, -0.01582876, -0.01572795, -0.01562715, -0.01552634, -0.01542553, -0.01532472, -0.01522392, -0.01512311, -0.0150223 , -0.0149215 , -0.01482069, -0.01472096, -0.01462126, -0.01452157, -0.01442187, -0.01432218, -0.01422248, -0.01412279, -0.01402309, -0.0139234 , -0.0138237 , -0.01372401, -0.01362431, -0.01352462, -0.01342492, -0.01332523, -0.01322553, -0.01312584, -0.01302615, -0.01292645, -0.01282739, -0.01272879, -0.01263019, -0.01253159, -0.01243298, -0.01233438, -0.01223578, -0.01213717, -0.01203857, -0.01193997, -0.01184137, -0.01174276, -0.01164416, -0.01154556, -0.01144695, -0.01134835, -0.01124975, -0.01115114, ... -0.00910467, -0.0090081 , -0.00891153, -0.00881496, -0.00871839, -0.00862181, -0.00852524, -0.00842867, -0.0083321 , -0.00823553, -0.00813895, -0.00804238, -0.00794581, -0.00784924, -0.00775302, -0.00765734, -0.00756166, -0.00746598, -0.0073703 , -0.00727461, -0.00717893, -0.00708325, -0.00698757, -0.00689188, -0.0067962 , -0.00670052, -0.00660484, -0.00650916, -0.00641347, -0.00631779, -0.00622251, -0.0061276 , -0.00603268, -0.00593777, -0.00584286, -0.00574795, -0.00565304, -0.00555813, -0.00546322, -0.00536831, -0.0052734 , -0.00517849, -0.00508358, -0.00498867, -0.00489376, -0.00479895, -0.00470467, -0.00461039, -0.00451611, -0.00442182, -0.00432754, -0.00423326, -0.00413898, -0.0040447 , -0.00395042, -0.00385614, -0.00376186, -0.00366758, -0.0035733 , -0.00347902, -0.00338483, -0.00329102, -0.0031972 , -0.00310339, -0.00300958, -0.00291576, -0.00282195, -0.00272813, -0.00263432, -0.0025405 , -0.00244669, -0.00235288, -0.00225906, -0.00216525, -0.00207143, -0.00197778, -0.00188425, -0.00179072, -0.00169719, -0.00160367, -0.00151014, -0.00141661, -0.00132308, -0.00122956, -0.00113603, -0.0010425 , -0.00094897, -0.00085545, -0.00076192, -0.00066839, -0.00057486, -0.00048134, -0.00038781, -0.00029428, -0.00020075, -0.00010723]) - rho0()float641.025e+03
- long_name :
- mean density used in Boussinesq approximation
- units :
- kilogram meter-3
array(1025.)
- hc()float6475.0
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
array(75.)
- nHIS()int323600
- long_name :
- number of time-steps between history records
array(3600, dtype=int32)
- ndefDIA()int320
- long_name :
- number of time-steps between the creation of diagnostic files
array(0, dtype=int32)
- el()float642.674e+06
- long_name :
- domain length in the ETA-direction
- units :
- meter
array(2674366.462091)
- angle()float644.106e-14
- long_name :
- angle between XI-axis and EAST
- units :
- radians
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- angle, scalar
array(4.10609047e-14)
- pm()float640.0001799
- long_name :
- curvilinear coordinate metric in XI
- units :
- meter-1
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- pm, scalar
array(0.00017986)
- Znudg()float640.0
- long_name :
- free-surface nudging/relaxation inverse time scale
- units :
- day-1
array(0.)
- spherical()int321
- long_name :
- grid type logical switch
- flag_values :
- [0 1]
- flag_meanings :
- Cartesian spherical
array(1, dtype=int32)
- ndefHIS()int320
- long_name :
- number of time-steps between the creation of history files
array(0, dtype=int32)
- Akv_bak()float641e-05
- long_name :
- background vertical mixing coefficient for momentum
- units :
- meter2 second-1
array(1.e-05)
- z_rho_v0(z_rho)float64-108.0 -107.5 ... -1.076 -0.5765
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array([-107.99277326, -107.49318161, -106.99358997, -106.49399832, -105.99440667, -105.49481502, -104.99522337, -104.49563173, -103.99604008, -103.49644843, -102.99685678, -102.49726514, -101.99767349, -101.49808184, -100.99849019, -100.49889854, -99.9993069 , -99.49971525, -99.0001236 , -98.50053195, -98.00093764, -97.50134204, -97.00174644, -96.50215084, -96.00255524, -95.50295964, -95.00336404, -94.50376843, -94.00417283, -93.50457723, -93.00498163, -92.50538603, -92.00579043, -91.50619483, -91.00659923, -90.50700363, -90.00740803, -89.50781243, -89.00821683, -88.50862122, -88.00902562, -87.50943002, -87.00983201, -86.5102324 , -86.01063279, -85.51103319, -85.01143358, -84.51183397, -84.01223437, -83.51263476, -83.01303516, -82.51343555, -82.01383594, -81.51423634, -81.01463673, -80.51503712, -80.01543752, -79.51583791, -79.0162383 , -78.5166387 , -78.01703909, -77.51743948, -77.01783988, -76.51823641, -76.0186328 , -75.5190292 , -75.01942559, -74.51982198, -74.02021838, -73.52061477, -73.02101116, -72.52140756, -72.02180395, -71.52220034, -71.02259674, -70.52299313, -70.02338952, -69.52378592, -69.02418231, -68.5245787 , ... -40.0467473 , -39.54712927, -39.04751123, -38.5478932 , -38.04827516, -37.54865713, -37.04903909, -36.54942105, -36.04980302, -35.55018498, -35.05056695, -34.55094891, -34.05133088, -33.55171284, -33.05209338, -32.55247257, -32.05285176, -31.55323095, -31.05361014, -30.55398933, -30.05436852, -29.55474771, -29.0551269 , -28.55550609, -28.05588528, -27.55626447, -27.05664366, -26.55702285, -26.05740204, -25.55778087, -25.0581578 , -24.55853472, -24.05891165, -23.55928857, -23.0596655 , -22.56004242, -22.06041935, -21.56079627, -21.0611732 , -20.56155012, -20.06192705, -19.56230398, -19.0626809 , -18.56305783, -18.0634344 , -17.56380965, -17.0641849 , -16.56456015, -16.06493539, -15.56531064, -15.06568589, -14.56606114, -14.06643638, -13.56681163, -13.06718688, -12.56756213, -12.06793738, -11.56831262, -11.06868787, -10.56906255, -10.06943677, -9.56981099, -9.0701852 , -8.57055942, -8.07093363, -7.57130785, -7.07168207, -6.57205628, -6.0724305 , -5.57280472, -5.07317893, -4.57355315, -4.07392736, -3.57430158, -3.0746758 , -2.57505001, -2.07542423, -1.57579845, -1.07617266, -0.57654688]) - nl_visc4()float641.24e+09
- long_name :
- nonlinear model biharmonic mixing coefficient for momentum
- units :
- meter4 second-1
array(1.24e+09)
- grid()int321
- cf_role :
- grid_topology
- topology_dimension :
- 2
- node_dimensions :
- xi_psi eta_psi
- face_dimensions :
- xi_rho: xi_psi (padding: both) eta_rho: eta_psi (padding: both)
- edge1_dimensions :
- xi_u: xi_psi eta_u: eta_psi (padding: both)
- edge2_dimensions :
- xi_v: xi_psi (padding: both) eta_v: eta_psi
- node_coordinates :
- lon_psi lat_psi
- face_coordinates :
- lon_rho lat_rho
- edge1_coordinates :
- lon_u lat_u
- edge2_coordinates :
- lon_v lat_v
- vertical_dimensions :
- s_rho: s_w (padding: none)
array(1, dtype=int32)
- z_rho_u0(z_rho)float64-108.2 -107.7 ... -1.078 -0.5774
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array([-108.16886878, -107.6684189 , -107.16796902, -106.66751913, -106.16706925, -105.66661937, -105.16616949, -104.66571961, -104.16526973, -103.66481984, -103.16436996, -102.66392008, -102.1634702 , -101.66302032, -101.16257044, -100.66212055, -100.16167067, -99.66122079, -99.16077091, -98.66032103, -98.15987465, -97.65942998, -97.1589853 , -96.65854062, -96.15809595, -95.65765127, -95.15720659, -94.65676192, -94.15631724, -93.65587256, -93.15542789, -92.65498321, -92.15453853, -91.65409386, -91.15364918, -90.6532045 , -90.15275983, -89.65231515, -89.15187047, -88.6514258 , -88.15098112, -87.65053644, -87.15009495, -86.64965555, -86.14921614, -85.64877674, -85.14833734, -84.64789794, -84.14745853, -83.64701913, -83.14657973, -82.64614033, -82.14570092, -81.64526152, -81.14482212, -80.64438272, -80.14394331, -79.64350391, -79.14306451, -78.64262511, -78.1421857 , -77.6417463 , -77.1413069 , -76.64087258, -76.14043845, -75.64000431, -75.13957018, -74.63913604, -74.13870191, -73.63826777, -73.13783364, -72.6373995 , -72.13696536, -71.63653123, -71.13609709, -70.63566296, -70.13522882, -69.63479469, -69.13436055, -68.63392642, ... -40.10974139, -39.60932625, -39.10891111, -38.60849597, -38.10808084, -37.6076657 , -37.10725056, -36.60683543, -36.10642029, -35.60600515, -35.10559002, -34.60517488, -34.10475974, -33.60434461, -33.10393134, -32.60351986, -32.10310838, -31.60269689, -31.10228541, -30.60187393, -30.10146245, -29.60105096, -29.10063948, -28.600228 , -28.09981651, -27.59940503, -27.09899355, -26.59858207, -26.09817058, -25.59775956, -25.09735106, -24.59694256, -24.09653406, -23.59612556, -23.09571706, -22.59530856, -22.09490005, -21.59449155, -21.09408305, -20.59367455, -20.09326605, -19.59285755, -19.09244904, -18.59204054, -18.0916325 , -17.59122621, -17.09081991, -16.59041362, -16.09000733, -15.58960104, -15.08919474, -14.58878845, -14.08838216, -13.58797586, -13.08756957, -12.58716328, -12.08675699, -11.58635069, -11.0859444 , -10.58553885, -10.08513392, -9.58472898, -9.08432405, -8.58391911, -8.08351418, -7.58310924, -7.08270431, -6.58229938, -6.08189444, -5.58148951, -5.08108457, -4.58067964, -4.0802747 , -3.57986977, -3.07946483, -2.5790599 , -2.07865496, -1.57825003, -1.0778451 , -0.57744016]) - nSTA()int3230
- long_name :
- number of time-steps between stations records
array(30, dtype=int32)
- M3nudg()float641.0
- long_name :
- 3D momentum nudging/relaxation inverse time scale
- units :
- day-1
array(1.)
- nRST()int323600
- long_name :
- number of time-steps between restart records
- cycle :
- only latest two records are maintained
array(3600, dtype=int32)
- Tcline()float6475.0
- long_name :
- S-coordinate surface/bottom layer width
- units :
- meter
array(75.)
- Lm2CLM()int320
- long_name :
- 2D momentum climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- Lm3PSH()int320
- long_name :
- 3D momentum push processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- mask_psi()float641.0
- long_name :
- mask on psi-points
- flag_values :
- [0. 1.]
- flag_meanings :
- land water
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- node
array(1.)
- Lm3CLM()int320
- long_name :
- 3D momentum climatology processing switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- lat_psi()float64-0.025
- long_name :
- latitude of PSI-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_psi, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array(-0.02499995)
- s_rho(z_rho)float64-0.2463 -0.2455 ... -0.001633
- long_name :
- S-coordinate at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- standard_name :
- ocean_s_coordinate_g1
- formula_terms :
- s: s_rho C: Cs_r eta: zeta depth: h depth_c: hc
- field :
- s_rho, scalar
- axis :
- Z
array([-0.2463117 , -0.24546767, -0.24462364, -0.24377961, -0.24293557, -0.24209154, -0.24124751, -0.24040348, -0.23955944, -0.23871541, -0.23787138, -0.23702735, -0.23618332, -0.23533928, -0.23449525, -0.23365122, -0.23280719, -0.23196316, -0.23111912, -0.23027509, -0.22938918, -0.22848302, -0.22757686, -0.2266707 , -0.22576454, -0.22485838, -0.22395223, -0.22304607, -0.22213991, -0.22123375, -0.22032759, -0.21942143, -0.21851527, -0.21760911, -0.21670295, -0.21579679, -0.21489063, -0.21398447, -0.21307831, -0.21217215, -0.21126599, -0.21035984, -0.20941572, -0.20844661, -0.2074775 , -0.20650838, -0.20553927, -0.20457016, -0.20360105, -0.20263193, -0.20166282, -0.20069371, -0.19972459, -0.19875548, -0.19778637, -0.19681726, -0.19584814, -0.19487903, -0.19390992, -0.19294081, -0.19197169, -0.19100258, -0.19003347, -0.18900366, -0.18797168, -0.1869397 , -0.18590772, -0.18487574, -0.18384376, -0.18281178, -0.1817798 , -0.18074782, -0.17971584, -0.17868386, -0.17765188, -0.1766199 , -0.17558792, -0.17455594, -0.17352395, -0.17249197, -0.17145999, -0.17042801, -0.16935993, -0.16826625, -0.16717257, -0.16607889, -0.16498522, -0.16389154, -0.16279786, -0.16170419, -0.16061051, -0.15951683, -0.15842315, -0.15732948, -0.1562358 , -0.15514212, -0.15404845, -0.15295477, -0.15186109, -0.15076741, ... -0.12643301, -0.12522454, -0.12401608, -0.12280762, -0.12159915, -0.12039069, -0.11918223, -0.11797376, -0.1167653 , -0.11555683, -0.11434837, -0.11313991, -0.11193144, -0.11072298, -0.10949432, -0.10823557, -0.10697683, -0.10571808, -0.10445934, -0.10320059, -0.10194185, -0.1006831 , -0.09942436, -0.09816561, -0.09690687, -0.09564812, -0.09438938, -0.09313063, -0.09187189, -0.09061314, -0.08933203, -0.08802967, -0.08672731, -0.08542495, -0.08412259, -0.08282023, -0.08151787, -0.08021551, -0.07891315, -0.07761079, -0.07630843, -0.07500607, -0.07370371, -0.07240135, -0.07109899, -0.06979108, -0.06845313, -0.06711519, -0.06577725, -0.0644393 , -0.06310136, -0.06176341, -0.06042547, -0.05908753, -0.05774958, -0.05641164, -0.05507369, -0.05373575, -0.05239781, -0.05105986, -0.04971644, -0.04835213, -0.04698782, -0.04562352, -0.04425921, -0.0428949 , -0.0415306 , -0.04016629, -0.03880198, -0.03743767, -0.03607337, -0.03470906, -0.03334475, -0.03198045, -0.03061614, -0.02924294, -0.02786242, -0.0264819 , -0.02510138, -0.02372086, -0.02234034, -0.02095982, -0.0195793 , -0.01819878, -0.01681826, -0.01543774, -0.01405722, -0.0126767 , -0.01129618, -0.00991566, -0.00853514, -0.00715462, -0.0057741 , -0.00439358, -0.00301306, -0.00163254]) - z_rho_u(ocean_time, z_rho)float64-107.8 -107.3 ... -0.777 -0.2766
- positive :
- up
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array([[-107.84644088, -107.34595268, -106.84546447, ..., -1.24765904, -0.74721578, -0.24677253], [-107.84254181, -107.34205314, -106.84156447, ..., -1.24366126, -0.74321753, -0.24277381], [-107.84221858, -107.34172987, -106.84124116, ..., -1.24332984, -0.74288608, -0.24244232], ..., [-107.86571292, -107.365227 , -106.86474108, ..., -1.267419 , -0.76697803, -0.26653706], [-107.86934622, -107.36886073, -106.86837525, ..., -1.27114428, -0.77070375, -0.27026321], [-107.87551058, -107.37502583, -106.87454107, ..., -1.27746471, -0.77702491, -0.27658511]]) - ocean_time(ocean_time)datetime64[ns]1985-10-02T03:00:00 ... 1985-10-...
- axis :
- T
- standard_name :
- time
array(['1985-10-02T03:00:00.000000000', '1985-10-02T09:00:00.000000000', '1985-10-02T15:00:00.000000000', '1985-10-02T21:00:00.000000000', '1985-10-03T03:00:00.000000000', '1985-10-03T09:00:00.000000000', '1985-10-03T15:00:00.000000000', '1985-10-03T21:00:00.000000000', '1985-10-04T03:00:00.000000000', '1985-10-04T09:00:00.000000000', '1985-10-04T15:00:00.000000000', '1985-10-04T21:00:00.000000000', '1985-10-05T03:00:00.000000000', '1985-10-05T09:00:00.000000000', '1985-10-05T15:00:00.000000000', '1985-10-05T21:00:00.000000000', '1985-10-06T03:00:00.000000000', '1985-10-06T09:00:00.000000000', '1985-10-06T15:00:00.000000000', '1985-10-06T21:00:00.000000000'], dtype='datetime64[ns]') - lat_rho()float641.45e-14
- long_name :
- latitude of RHO-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_rho, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- axis :
- Y
array(1.44996862e-14)
- LuvSponge()int320
- long_name :
- horizontal viscosity sponge activation switch
- flag_values :
- [0 1]
- flag_meanings :
- .FALSE. .TRUE.
array(0, dtype=int32)
- nAVG()int32180
- long_name :
- number of time-steps between time-averaged records
array(180, dtype=int32)
- lat_v()float64-0.025
- long_name :
- latitude of V-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_v, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- axis :
- Y
array(-0.02499997)
- h()float644.314e+03
- long_name :
- bathymetry at RHO-points
- units :
- meter
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
- location :
- face
- field :
- bath, scalar
array(4313.57672913)
- z_rho_psi0(z_rho)float64-108.2 -107.7 ... -1.078 -0.5774
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array([-108.16789118, -107.66744606, -107.16700094, -106.66655583, -106.16611071, -105.66566559, -105.16522048, -104.66477536, -104.16433024, -103.66388512, -103.16344001, -102.66299489, -102.16254977, -101.66210466, -101.16165954, -100.66121442, -100.1607693 , -99.66032419, -99.15987907, -98.65943395, -98.15899231, -97.65855235, -97.15811238, -96.65767242, -96.15723246, -95.6567925 , -95.15635253, -94.65591257, -94.15547261, -93.65503264, -93.15459268, -92.65415272, -92.15371275, -91.65327279, -91.15283283, -90.65239287, -90.1519529 , -89.65151294, -89.15107298, -88.65063301, -88.15019305, -87.64975309, -87.14931627, -86.64888153, -86.14844679, -85.64801205, -85.14757731, -84.64714257, -84.14670783, -83.64627309, -83.14583835, -82.64540361, -82.14496887, -81.64453413, -81.14409939, -80.64366465, -80.14322991, -79.64279517, -79.14236043, -78.64192569, -78.14149095, -77.64105621, -77.14062147, -76.64019176, -76.13976224, -75.63933272, -75.13890319, -74.63847367, -74.13804414, -73.63761462, -73.13718509, -72.63675557, -72.13632604, -71.63589652, -71.13546699, -70.63503747, -70.13460794, -69.63417842, -69.1337489 , -68.63331937, ... -40.10939167, -39.60898096, -39.10857025, -38.60815954, -38.10774882, -37.60733811, -37.1069274 , -36.60651669, -36.10610598, -35.60569527, -35.10528455, -34.60487384, -34.10446313, -33.60405242, -33.10364356, -32.60323647, -32.10282938, -31.60242228, -31.10201519, -30.6016081 , -30.101201 , -29.60079391, -29.10038681, -28.59997972, -28.09957263, -27.59916553, -27.09875844, -26.59835135, -26.09794425, -25.59753762, -25.09713348, -24.59672934, -24.0963252 , -23.59592106, -23.09551692, -22.59511277, -22.09470863, -21.59430449, -21.09390035, -20.59349621, -20.09309207, -19.59268793, -19.09228379, -18.59187964, -18.09147596, -17.591074 , -17.09067205, -16.5902701 , -16.08986814, -15.58946619, -15.08906423, -14.58866228, -14.08826032, -13.58785837, -13.08745642, -12.58705446, -12.08665251, -11.58625055, -11.0858486 , -10.58544738, -10.08504677, -9.58464617, -9.08424556, -8.58384495, -8.08344434, -7.58304373, -7.08264312, -6.58224251, -6.0818419 , -5.58144129, -5.08104068, -4.58064007, -4.08023947, -3.57983886, -3.07943825, -2.57903764, -2.07863703, -1.57823642, -1.07783581, -0.5774352 ]) - lon_psi()float64-140.0
- long_name :
- longitude of PSI-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_psi, scalar
- grid :
- <xgcm.Grid> X Axis (not periodic): * center xi_rho --> inner * inner xi_u --> center Y Axis (not periodic): * center eta_rho --> inner * inner eta_v --> center Z Axis (not periodic): * center s_rho --> outer * outer s_w --> center
array(-140.025)
- Vstretching()int321
- long_name :
- vertical terrain-following stretching function
array(1, dtype=int32)
- u(ocean_time, z_rho)float640.0935 0.09244 ... 0.1627 0.1625
- file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- format :
- netCDF-3 64bit offset file
- Conventions :
- CF-1.4, SGRID-0.3
- type :
- ROMS/TOMS diagnostics file
- title :
- TPOS 1/20deg
- var_info :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/varinfo.dat
- rst_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2_2.nc
- avg_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2_2.nc
- dia_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- sta_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2_2.nc
- grd_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_grd.nc
- ini_file :
- /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
- frc_file_01 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_ncep.nc
- frc_file_02 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_giss.nc
- frc_file_03 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_gcgcs.nc
- bry_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_bry.nc
- script_file :
- ./tpos20_3_diag_2.in
- spos_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/stations_tpos20.in
- NLM_LBC :
- EDGE: WEST SOUTH EAST NORTH zeta: Che Che Che Che ubar: Shc Shc Shc Shc vbar: Shc Shc Shc Shc u: RadNud RadNud RadNud RadNud v: RadNud RadNud RadNud RadNud temp: RadNud RadNud Cla RadNud salt: RadNud RadNud Cla RadNud
- svn_url :
- https:://myroms.org/svn/src
- svn_rev :
- Unversioned directory
- code_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROMS/ROMS
- header_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INC/
- header_file :
- tpos20.h
- os :
- Linux
- cpu :
- x86_64
- compiler_system :
- ifort
- compiler_command :
- /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/mpi/mpif90
- compiler_flags :
- -fp-model precise -ip -O3
- tiling :
- 018x012
- history :
- ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:53:43 PM
- ana_file :
- ROMS/Functionals/ana_btflux.h
- CPP_options :
- TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SHAPE, AVERAGES, !BOUNDARY_A BULK_FLUXES, CCSM_FLUXES, !COLLECT_ALL..., CURVGRID, DIAGNOSTICS_TS, DIAGNOSTICS_UV, DJ_GRADPS, DOUBLE_PRECISION, EMINUSP, INLINE_2DIO, LMD_CONVEC, LMD_DDMIX, LMD_MIXING, LMD_NONLOCAL, LMD_RIMIX, LMD_SHAPIRO, LMD_SKPP, LONGWAVE_OUT, MASKING, MIX_ISO_TS, MIX_S_UV, MPI, NONLINEAR, NONLIN_EOS, NO_LBC_ATT, PERFECT_RESTART, POWER_LAW, PROFILE, RADIATION_2D, REDUCE_ALLGATHER, RI_SPLINES, !RST_SINGLE, SALINITY, SOLAR_SOURCE, SOLVE3D, SPLINES_VDIFF, SPLINES_VVISC, STATIONS, TS_U3HADVECTION, TS_C4VADVECTION, TS_DIF4, UV_ADV, UV_COR, UV_U3HADVECTION, UV_C4VADVECTION, UV_QDRAG, UV_VIS4, VAR_RHO_2D
- description :
- sum over ['u_prsgrd', 'u_hadv', 'u_vadv', 'u_hvisc']
array([[ 0.0934992 , 0.0924413 , 0.09138341, ..., 0.11522851, 0.11497532, 0.11472214], [ 0.07715398, 0.07701738, 0.07688077, ..., 0.13610994, 0.13541674, 0.13472353], [-0.07029411, -0.06939658, -0.06849905, ..., 0.01532662, 0.01492513, 0.01452364], ..., [ 0.04566397, 0.04639652, 0.04712908, ..., 0.05993552, 0.05920398, 0.05847243], [ 0.05014986, 0.05335076, 0.05655166, ..., 0.06999024, 0.06922207, 0.06845391], [ 0.14647725, 0.14917367, 0.15187009, ..., 0.1629429 , 0.16272458, 0.16250625]]) - v(ocean_time, z_rho)float640.106 0.1083 ... -0.0536 -0.05361
- file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- format :
- netCDF-3 64bit offset file
- Conventions :
- CF-1.4, SGRID-0.3
- type :
- ROMS/TOMS diagnostics file
- title :
- TPOS 1/20deg
- var_info :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/varinfo.dat
- rst_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2_2.nc
- avg_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2_2.nc
- dia_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- sta_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2_2.nc
- grd_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_grd.nc
- ini_file :
- /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
- frc_file_01 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_ncep.nc
- frc_file_02 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_giss.nc
- frc_file_03 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_gcgcs.nc
- bry_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_bry.nc
- script_file :
- ./tpos20_3_diag_2.in
- spos_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/stations_tpos20.in
- NLM_LBC :
- EDGE: WEST SOUTH EAST NORTH zeta: Che Che Che Che ubar: Shc Shc Shc Shc vbar: Shc Shc Shc Shc u: RadNud RadNud RadNud RadNud v: RadNud RadNud RadNud RadNud temp: RadNud RadNud Cla RadNud salt: RadNud RadNud Cla RadNud
- svn_url :
- https:://myroms.org/svn/src
- svn_rev :
- Unversioned directory
- code_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROMS/ROMS
- header_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INC/
- header_file :
- tpos20.h
- os :
- Linux
- cpu :
- x86_64
- compiler_system :
- ifort
- compiler_command :
- /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/mpi/mpif90
- compiler_flags :
- -fp-model precise -ip -O3
- tiling :
- 018x012
- history :
- ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:53:43 PM
- ana_file :
- ROMS/Functionals/ana_btflux.h
- CPP_options :
- TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SHAPE, AVERAGES, !BOUNDARY_A BULK_FLUXES, CCSM_FLUXES, !COLLECT_ALL..., CURVGRID, DIAGNOSTICS_TS, DIAGNOSTICS_UV, DJ_GRADPS, DOUBLE_PRECISION, EMINUSP, INLINE_2DIO, LMD_CONVEC, LMD_DDMIX, LMD_MIXING, LMD_NONLOCAL, LMD_RIMIX, LMD_SHAPIRO, LMD_SKPP, LONGWAVE_OUT, MASKING, MIX_ISO_TS, MIX_S_UV, MPI, NONLINEAR, NONLIN_EOS, NO_LBC_ATT, PERFECT_RESTART, POWER_LAW, PROFILE, RADIATION_2D, REDUCE_ALLGATHER, RI_SPLINES, !RST_SINGLE, SALINITY, SOLAR_SOURCE, SOLVE3D, SPLINES_VDIFF, SPLINES_VVISC, STATIONS, TS_U3HADVECTION, TS_C4VADVECTION, TS_DIF4, UV_ADV, UV_COR, UV_U3HADVECTION, UV_C4VADVECTION, UV_QDRAG, UV_VIS4, VAR_RHO_2D
- description :
- sum over ['v_prsgrd', 'v_hadv', 'v_vadv', 'v_hvisc']
array([[ 0.10599922, 0.10827727, 0.11055531, ..., 0.0872403 , 0.08617826, 0.08511622], [ 0.02181563, 0.02367078, 0.02552593, ..., -0.03674249, -0.03789036, -0.03903822], [ 0.09048007, 0.09216873, 0.09385739, ..., 0.0878034 , 0.08692331, 0.08604321], ..., [-0.03156965, -0.02900752, -0.02644538, ..., 0.03970313, 0.03975124, 0.03979936], [-0.16548416, -0.16360047, -0.16171677, ..., 0.00489039, 0.00489994, 0.00490948], [-0.11667727, -0.11486929, -0.11306131, ..., -0.05359522, -0.05360288, -0.05361054]]) - temp(ocean_time, z_rho)float64-0.8086 -0.7765 ... -0.02899
- file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- format :
- netCDF-3 64bit offset file
- Conventions :
- CF-1.4, SGRID-0.3
- type :
- ROMS/TOMS diagnostics file
- title :
- TPOS 1/20deg
- var_info :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/varinfo.dat
- rst_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2_2.nc
- avg_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2_2.nc
- dia_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- sta_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2_2.nc
- grd_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_grd.nc
- ini_file :
- /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
- frc_file_01 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_ncep.nc
- frc_file_02 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_giss.nc
- frc_file_03 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_gcgcs.nc
- bry_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_bry.nc
- script_file :
- ./tpos20_3_diag_2.in
- spos_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/stations_tpos20.in
- NLM_LBC :
- EDGE: WEST SOUTH EAST NORTH zeta: Che Che Che Che ubar: Shc Shc Shc Shc vbar: Shc Shc Shc Shc u: RadNud RadNud RadNud RadNud v: RadNud RadNud RadNud RadNud temp: RadNud RadNud Cla RadNud salt: RadNud RadNud Cla RadNud
- svn_url :
- https:://myroms.org/svn/src
- svn_rev :
- Unversioned directory
- code_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROMS/ROMS
- header_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INC/
- header_file :
- tpos20.h
- os :
- Linux
- cpu :
- x86_64
- compiler_system :
- ifort
- compiler_command :
- /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/mpi/mpif90
- compiler_flags :
- -fp-model precise -ip -O3
- tiling :
- 018x012
- history :
- ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:53:43 PM
- ana_file :
- ROMS/Functionals/ana_btflux.h
- CPP_options :
- TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SHAPE, AVERAGES, !BOUNDARY_A BULK_FLUXES, CCSM_FLUXES, !COLLECT_ALL..., CURVGRID, DIAGNOSTICS_TS, DIAGNOSTICS_UV, DJ_GRADPS, DOUBLE_PRECISION, EMINUSP, INLINE_2DIO, LMD_CONVEC, LMD_DDMIX, LMD_MIXING, LMD_NONLOCAL, LMD_RIMIX, LMD_SHAPIRO, LMD_SKPP, LONGWAVE_OUT, MASKING, MIX_ISO_TS, MIX_S_UV, MPI, NONLINEAR, NONLIN_EOS, NO_LBC_ATT, PERFECT_RESTART, POWER_LAW, PROFILE, RADIATION_2D, REDUCE_ALLGATHER, RI_SPLINES, !RST_SINGLE, SALINITY, SOLAR_SOURCE, SOLVE3D, SPLINES_VDIFF, SPLINES_VVISC, STATIONS, TS_U3HADVECTION, TS_C4VADVECTION, TS_DIF4, UV_ADV, UV_COR, UV_U3HADVECTION, UV_C4VADVECTION, UV_QDRAG, UV_VIS4, VAR_RHO_2D
- description :
- sum over ['temp_hadv', 'temp_vadv', 'temp_hdiff']
array([[-0.80857886, -0.77648844, -0.74439801, ..., 0.18671655, 0.18685957, 0.18700259], [-0.26256931, -0.24553172, -0.22849412, ..., 0.14542021, 0.14556302, 0.14570583], [ 0.83157955, 0.80610695, 0.78063435, ..., 0.10669244, 0.10677824, 0.10686404], ..., [-1.21461383, -1.1704578 , -1.12630177, ..., 0.0135132 , 0.01434176, 0.01517032], [-1.48031712, -1.44996477, -1.41961242, ..., -0.00747166, -0.00707024, -0.00666881], [-0.68760617, -0.67779509, -0.66798401, ..., -0.02890075, -0.02894501, -0.02898926]]) - salt(ocean_time, z_rho)float64-0.02245 -0.02013 ... 0.0006571
- file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- format :
- netCDF-3 64bit offset file
- Conventions :
- CF-1.4, SGRID-0.3
- type :
- ROMS/TOMS diagnostics file
- title :
- TPOS 1/20deg
- var_info :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/varinfo.dat
- rst_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_rst_tpos20_3_d2_2.nc
- avg_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_avg_tpos20_3_d2_2.nc
- dia_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_dia_tpos20_3_d2_2.nc
- sta_file :
- /glade/scratch/dwhitt/tpos20/OUT/ocean_sta_tpos20_3_d2_2.nc
- grd_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_grd.nc
- ini_file :
- /glade/p/nsc/ncgd0043/tpos20/OUT/ocean_rst_tpos20_3_d2.nc
- frc_file_01 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_ncep.nc
- frc_file_02 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_giss.nc
- frc_file_03 :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/FRC/ocean_frc_gcgcs.nc
- bry_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/ocean_bry.nc
- script_file :
- ./tpos20_3_diag_2.in
- spos_file :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INI/stations_tpos20.in
- NLM_LBC :
- EDGE: WEST SOUTH EAST NORTH zeta: Che Che Che Che ubar: Shc Shc Shc Shc vbar: Shc Shc Shc Shc u: RadNud RadNud RadNud RadNud v: RadNud RadNud RadNud RadNud temp: RadNud RadNud Cla RadNud salt: RadNud RadNud Cla RadNud
- svn_url :
- https:://myroms.org/svn/src
- svn_rev :
- Unversioned directory
- code_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/ROMS/ROMS
- header_dir :
- /glade/p/nsc/ncgd0043/tpos_ROMS_runs_setup_scripts/tpos20/INC/
- header_file :
- tpos20.h
- os :
- Linux
- cpu :
- x86_64
- compiler_system :
- ifort
- compiler_command :
- /glade/u/apps/ch/opt/ncarcompilers/0.5.0/intel/18.0.5/mpi/mpif90
- compiler_flags :
- -fp-model precise -ip -O3
- tiling :
- 018x012
- history :
- ROMS/TOMS, Version 3.7, Tuesday - July 30, 2019 - 5:53:43 PM
- ana_file :
- ROMS/Functionals/ana_btflux.h
- CPP_options :
- TPOS20, ALBEDO_JRA, ANA_BSFLUX, ANA_BTFLUX, ASSUMED_SHAPE, AVERAGES, !BOUNDARY_A BULK_FLUXES, CCSM_FLUXES, !COLLECT_ALL..., CURVGRID, DIAGNOSTICS_TS, DIAGNOSTICS_UV, DJ_GRADPS, DOUBLE_PRECISION, EMINUSP, INLINE_2DIO, LMD_CONVEC, LMD_DDMIX, LMD_MIXING, LMD_NONLOCAL, LMD_RIMIX, LMD_SHAPIRO, LMD_SKPP, LONGWAVE_OUT, MASKING, MIX_ISO_TS, MIX_S_UV, MPI, NONLINEAR, NONLIN_EOS, NO_LBC_ATT, PERFECT_RESTART, POWER_LAW, PROFILE, RADIATION_2D, REDUCE_ALLGATHER, RI_SPLINES, !RST_SINGLE, SALINITY, SOLAR_SOURCE, SOLVE3D, SPLINES_VDIFF, SPLINES_VVISC, STATIONS, TS_U3HADVECTION, TS_C4VADVECTION, TS_DIF4, UV_ADV, UV_COR, UV_U3HADVECTION, UV_C4VADVECTION, UV_QDRAG, UV_VIS4, VAR_RHO_2D
- description :
- sum over ['salt_hadv', 'salt_vadv', 'salt_hdiff']
array([[-0.02245094, -0.02013102, -0.0178111 , ..., -0.02703016, -0.02698611, -0.02694206], [-0.01065129, -0.00779557, -0.00493986, ..., -0.02448466, -0.02449072, -0.02449678], [ 0.04670271, 0.04672056, 0.04673841, ..., -0.02254857, -0.02255921, -0.02256986], ..., [-0.10480203, -0.0995391 , -0.09427616, ..., -0.00719575, -0.00735464, -0.00751353], [-0.17697424, -0.17046454, -0.16395484, ..., -0.00594357, -0.00615052, -0.00635747], [-0.16151997, -0.15618638, -0.15085279, ..., 0.0005837 , 0.00062042, 0.00065713]])
write_to_txt(
rhs.rename(renamer).pipe(interleave_time, t0),
outdir=outdir,
prefix="ocean_colpsh_",
)
initial condition#
substate = (subds[["U", "W", "T", "S"]].compute()) - offset
ic = substate.cf.sel(time=t0)
write_to_txt(ic.compute(), outdir=outdir, prefix="ocean_ic_")
restoring#
write_to_txt(interleave_time(substate, t0), outdir=outdir, prefix="ocean_colfrc_")
surface forcing#
frc = subds[["sustr", "svstr", "swrad", "ssflux"]]
frc["shfluxminusswrad"] = subds["shflux"] - frc["swrad"]
time = subds.indexes["ocean_time"].to_numpy()
frc["time"] = ("ocean_time", (time - t0).astype("timedelta64[s]").astype(float))
frc.load()
write_to_txt(frc, outdir=outdir, prefix="ocean_frc_")
# Test flux calculations
shflux2 = subds[["lwrad", "latent", "sensible"]].to_array().sum("variable")
subds.swrad.plot()
subds.shflux.plot()
(-subds.swrad + subds.shflux).plot(marker="x")
(shflux2).plot()
[<matplotlib.lines.Line2D at 0x2b245c327940>]