miχpods#
test out hvplot
build LES catalog
bootstrap error on mean, median?
switch to daily data?
add TAO N2, Tz, S2
add TAO χpods
move enso_transition_mask to mixpods
fix EUC max at 110
References#
Warner & Moum (2019)
Setup#
Show code cell source
%load_ext watermark
import os
import cf_xarray
import dask
import dcpy
import distributed
import flox.xarray
import hvplot.xarray
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
import pump
from pump import mixpods
dask.config.set({"array.slicing.split_large_chunks": False})
mpl.rcParams["figure.dpi"] = 140
xr.set_options(keep_attrs=True)
gcmdir = "/glade/campaign/cgd/oce/people/bachman/TPOS_1_20_20_year/OUTPUT/" # MITgcm output directory
stationdirname = gcmdir
%watermark -iv
Show code cell output
hvplot : 0.8.0
dcpy : 0.1.dev385+g121534c
pump : 0.1
numpy : 1.22.4
sys : 3.10.5 | packaged by conda-forge | (main, Jun 14 2022, 07:04:59) [GCC 10.3.0]
distributed: 2022.7.0
json : 2.0.9
matplotlib : 3.5.2
cf_xarray : 0.7.3
xarray : 2022.6.0rc0
dask : 2022.7.0
flox : 0.5.9
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask_jobqueue/core.py:20: FutureWarning: tmpfile is deprecated and will be removed in a future release. Please use dask.utils.tmpfile instead.
from distributed.utils import tmpfile
Show code cell source
import ncar_jobqueue
if "client" in locals():
client.close()
del client
if "cluster" in locals():
cluster.close()
# env = {"OMP_NUM_THREADS": "3", "NUMBA_NUM_THREADS": "3"}
# cluster = distributed.LocalCluster(
# n_workers=8,
# threads_per_worker=1,
# env=env
# )
if "cluster" in locals():
del cluster
# cluster = ncar_jobqueue.NCARCluster(
# project="NCGD0011",
# scheduler_options=dict(dashboard_address=":9797"),
# )
# cluster = dask_jobqueue.PBSCluster(
# cores=9, processes=9, memory="108GB", walltime="02:00:00", project="NCGD0043",
# env_extra=env,
# )
import dask_jobqueue
cluster = dask_jobqueue.PBSCluster(
cores=1, # The number of cores you want
memory="23GB", # Amount of memory
processes=1, # How many processes
queue="casper", # The type of queue to utilize (/glade/u/apps/dav/opt/usr/bin/execcasper)
local_directory="$TMPDIR", # Use your local directory
resource_spec="select=1:ncpus=1:mem=23GB", # Specify resources
project="ncgd0011", # Input your project ID here
walltime="02:00:00", # Amount of wall time
interface="ib0", # Interface to use
)
cluster.scale(jobs=4)
Show code cell source
client = distributed.Client(cluster)
client
Show code cell output
Client
Client-b2e75700-02ce-11ed-b1d0-3cecef1acbfa
| Connection method: Cluster object | Cluster type: dask_jobqueue.PBSCluster |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/dcherian/proxy/8787/status |
Cluster Info
PBSCluster
e67d7989
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/dcherian/proxy/8787/status | Workers: 0 |
| Total threads: 0 | Total memory: 0 B |
Scheduler Info
Scheduler
Scheduler-b839cee5-f0db-4274-abbd-4e85bfea61cc
| Comm: tcp://10.12.206.63:36269 | Workers: 0 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/dcherian/proxy/8787/status | Total threads: 0 |
| Started: Just now | Total memory: 0 B |
Workers
tao_gridded = xr.open_dataset(
os.path.expanduser("~/work/pump/zarrs/tao-gridded-ancillary.zarr"),
chunks="auto",
engine="zarr",
).sel(longitude=-140, time=slice("2005-Jun", "2015"))
tao_gridded["depth"].attrs["axis"] = "Z"
# eucmax exists
tao_gridded.coords["eucmax"] = pump.calc.get_euc_max(
tao_gridded.u.reset_coords(drop=True), kind="data"
)
# pump.calc.calc_reduced_shear(tao_gridded)
tao_gridded.coords["enso_transition"] = pump.obs.make_enso_transition_mask().reindex(
time=tao_gridded.time, method="nearest"
)
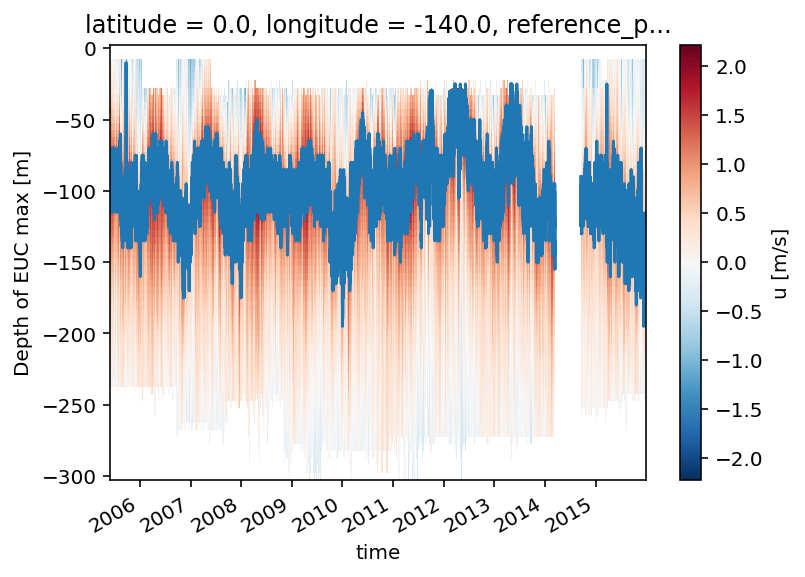
tao_gridded.u.cf.plot()
tao_gridded.eucmax.plot()
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/dataset.py:248: UserWarning: The specified Dask chunks separate the stored chunks along dimension "depth" starting at index 58. This could degrade performance. Instead, consider rechunking after loading.
warnings.warn(
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/dataset.py:248: UserWarning: The specified Dask chunks separate the stored chunks along dimension "time" starting at index 139586. This could degrade performance. Instead, consider rechunking after loading.
warnings.warn(
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/dataset.py:248: UserWarning: The specified Dask chunks separate the stored chunks along dimension "longitude" starting at index 2. This could degrade performance. Instead, consider rechunking after loading.
warnings.warn(
[<matplotlib.lines.Line2D at 0x2adc519719c0>]
tao_gridded = tao_gridded.update(
{
"n2s2pdf": mixpods.pdf_N2S2(
tao_gridded[["S2", "N2T"]]
.drop_vars(["shallowest", "zeuc"])
.rename_vars({"N2T": "N2"})
).load()
}
)
tao_Ri = xr.load_dataarray(
"tao-hourly-Ri-seasonal-percentiles.nc"
).cf.guess_coord_axis()
metrics = pump.model.read_metrics(stationdirname)
stations = pump.model.read_stations_20(stationdirname)
stations
<xarray.Dataset>
Dimensions: (depth: 185, time: 174000, longitude: 12, latitude: 111)
Coordinates:
* depth (depth) float32 -1.25 -3.75 -6.25 ... -5.658e+03 -5.758e+03
* latitude (latitude) float64 -3.075 -3.025 -2.975 ... 5.925 5.975 6.025
* longitude (longitude) float64 -155.1 -155.0 -155.0 ... -110.0 -110.0
* time (time) datetime64[ns] 1998-12-31T18:00:00 ... 2018-11-06T1...
Data variables: (12/20)
DFrI_TH (depth, time, longitude, latitude) float32 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
KPPdiffKzT (depth, time, longitude, latitude) float32 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
KPPg_TH (depth, time, longitude, latitude) float32 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
KPPhbl (time, longitude, latitude) float32 dask.array<chunksize=(6000, 3, 3), meta=np.ndarray>
KPPviscAz (depth, time, longitude, latitude) float32 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
SSH (time, longitude, latitude) float32 dask.array<chunksize=(6000, 3, 3), meta=np.ndarray>
... ...
nonlocal_flux (depth, time, longitude, latitude) float64 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
dens (depth, time, longitude, latitude) float32 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
mld (time, longitude, latitude) float32 dask.array<chunksize=(6000, 3, 3), meta=np.ndarray>
Jq (depth, time, longitude, latitude) float64 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
dJdz (depth, time, longitude, latitude) float64 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
dTdt (depth, time, longitude, latitude) float64 dask.array<chunksize=(185, 6000, 3, 3), meta=np.ndarray>
Attributes:
easting: longitude
northing: latitude
title: Station profile, index (i,j)=(1201,240)gcmeq = stations.sel(
latitude=0, longitude=[-155.025, -140.025, -125.025, -110.025], method="nearest"
)
# enso = pump.obs.make_enso_mask()
# mitgcm["enso"] = enso.reindex(time=mitgcm.time.data, method="nearest")
gcmeq["eucmax"] = pump.calc.get_euc_max(gcmeq.u)
pump.calc.calc_reduced_shear(gcmeq)
gcmeq["enso_transition"] = pump.obs.make_enso_transition_mask().reindex(
time=gcmeq.time.data, method="nearest"
)
mitgcm = gcmeq.sel(longitude=-140.025, method="nearest")
# metrics_ = xr.align(metrics, mitgcm.expand_dims(["latitude", "longitude"]), join="inner")[0].squeeze()
mitgcm = mitgcm.update({"n2s2pdf": mixpods.pdf_N2S2(mitgcm.load())})
calc uz
calc vz
calc S2
calc N2
calc shred2
Calc Ri
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/computation.py:771: RuntimeWarning: divide by zero encountered in log10
result_data = func(*input_data)
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/computation.py:771: RuntimeWarning: invalid value encountered in log10
result_data = func(*input_data)
gcmeq.eucmax.hvplot.line(by="longitude")