miχpods for LES simulations#
import ast
import os
import dcpy
import intake
import matplotlib as mpl
import matplotlib.pyplot as plt
import xarray as xr
import xgcm
from datatree import DataTree
import hvplot.xarray
import pump
from pump import mixpods
plt.rcParams["figure.dpi"] = 140
import holoviews as hv
hv.notebook_extension("bokeh")
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask_jobqueue/core.py:20: FutureWarning: tmpfile is deprecated and will be removed in a future release. Please use dask.utils.tmpfile instead.
from distributed.utils import tmpfile
Read data#
LES#
les_catalog = intake.open_esm_datastore(
"../catalogs/pump-les-catalog.json",
read_csv_kwargs={"converters": {"variables": ast.literal_eval}},
)
les_catalog.df
| length | kind | longitude | latitude | month | path | variables | |
|---|---|---|---|---|---|---|---|
| 0 | 5-day | average | -120 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 1 | 5-day | average | -165 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 2 | 5-day | average | -115 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 3 | 5-day | average | -110 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 4 | 5-day | average | -105 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 5 | 5-day | average | -100 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 6 | 5-day | average | -160 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 7 | 5-day | average | -155 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 8 | 5-day | average | -150 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 9 | 5-day | average | -145 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 10 | 5-day | average | -140 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 11 | 5-day | average | -135 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 12 | 5-day | average | -130 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 13 | 5-day | average | -125 | 0.0 | may | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 14 | month | average | -140 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 15 | month | mooring | -140 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [u, v, w, temp, salt, nu_sgs, kappa_sgs, alpha... |
| 16 | month | average | -140 | 1.5 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 17 | month | mooring | -140 | 1.5 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [u, v, w, temp, salt, nu_sgs, kappa_sgs, alpha... |
| 18 | month | average | -140 | -1.5 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 19 | month | mooring | -140 | -1.5 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [u, v, w, temp, salt, nu_sgs, kappa_sgs, alpha... |
| 20 | 5-day | average | -120 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 21 | 5-day | average | -165 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 22 | 5-day | average | -115 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 23 | 5-day | average | -110 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 24 | 5-day | average | -105 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 25 | 5-day | average | -100 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 26 | 5-day | average | -160 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 27 | 5-day | average | -155 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 28 | 5-day | average | -150 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 29 | 5-day | average | -145 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 30 | 5-day | average | -135 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 31 | 5-day | average | -130 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 32 | 5-day | average | -125 | 0.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 33 | month | average | -140 | 3.0 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 34 | month | average | -140 | 4.5 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [ume, vme, tempme, saltme, urms, vrms, wrms, t... |
| 35 | month | mooring | -140 | 4.5 | oct | /glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_r... | [u, v, w, temp, salt, nu_sgs, kappa_sgs, alpha... |
Only month-long “moorings”
mooring_datasets = les_catalog.search(kind="mooring", length="month").to_dataset_dict(
preprocess=pump.les.preprocess_les_dataset
)
moorings = DataTree.from_dict(mooring_datasets).squeeze()
--> The keys in the returned dictionary of datasets are constructed as follows:
'latitude.longitude.month.kind.length'
100.00% [4/4 00:00<00:00]
avg_datasets = catalog.search(kind="average", length="month").to_dataset_dict(
preprocess=pump.les.preprocess_les_dataset
)
avgs = DataTree.from_dict(avg_datasets).squeeze()
avgs = avgs.rename_vars({"ume": "u", "vme": "v"})les_
--> The keys in the returned dictionary of datasets are constructed as follows:
'latitude.longitude.month.kind.length'
100.00% [5/5 00:10<00:00]
def clear(dataset):
new = dataset.copy()
for var in dataset.variables:
del new[var]
return new
def clear_root(tree):
new = tree.copy()
for var in tree.ds.variables:
del new.ds[var]
return new
def extract(tree, varnames):
return tree.map_over_subtree(lambda ds: ds[varnames])
def to_dataset(tree, dim):
return xr.concat(
[
child.ds.expand_dims({dim: [name]} if dim not in child.ds else dim)
for name, child in tree.children.items()
],
dim=dim,
)
def add_ancillary_mixpod_variables(tree):
from datatree import DataTree
tree = clear_root(tree)
# grid = xgcm.Grid(
# avgs["0.0.-140.oct.average.month"].ds,
# coords={"Z": {"center": "z", "inner": "zc"}},
# metrics={("Z",): "dz"},
# )
tree.map_over_subtree_inplace(mixpods.prepare)
tree = tree.assign({"n2s2pdf": mixpods.pdf_N2S2})
tree["n2s2pdf"] = (
to_dataset(extract(tree, "n2s2pdf"), dim="latitude")
.sortby("latitude")
.to_array()
.squeeze("variable")
.load()
)
return tree
moorings = add_ancillary_mixpod_variables(moorings)
avgs = add_ancillary_mixpod_variables(avgs)
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
Read TAO#
tao_gridded = xr.open_dataset(
os.path.expanduser("~/work/pump/zarrs/tao-gridded-ancillary.zarr"),
chunks="auto",
engine="zarr",
).sel(longitude=-140, time=slice("2005-Jun", "2015"))
tao_gridded["depth"].attrs["axis"] = "Z"
# eucmax exists
tao_gridded.coords["eucmax"] = pump.calc.get_euc_max(
tao_gridded.u.reset_coords(drop=True), kind="data"
)
# pump.calc.calc_reduced_shear(tao_gridded)
tao_gridded.coords["enso_transition"] = pump.obs.make_enso_transition_mask().reindex(
time=tao_gridded.time, method="nearest"
)
tao_gridded = tao_gridded.update(
{
"n2s2pdf": mixpods.pdf_N2S2(
tao_gridded[["S2", "N2T"]]
.drop_vars(["shallowest", "zeuc"])
.rename_vars({"N2T": "N2"})
).load()
}
)
ds = tao_gridded[["N2", "S2"]].drop_vars(["shallowest", "zeuc"])
import flox.xarray
import numpy as np
tao_gridded["n2s2pdf_monthly"] = mixpods.to_density(
flox.xarray.xarray_reduce(
ds.S2,
np.log10(4 * ds.N2),
np.log10(ds.S2),
ds.time.dt.month,
func="count",
expected_groups=(np.linspace(-5, -2, 30), np.linspace(-5, -2, 30), None),
isbin=(True, True, False),
).load()
)
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/dataset.py:248: UserWarning: The specified Dask chunks separate the stored chunks along dimension "depth" starting at index 58. This could degrade performance. Instead, consider rechunking after loading.
warnings.warn(
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/dataset.py:248: UserWarning: The specified Dask chunks separate the stored chunks along dimension "time" starting at index 139586. This could degrade performance. Instead, consider rechunking after loading.
warnings.warn(
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/dataset.py:248: UserWarning: The specified Dask chunks separate the stored chunks along dimension "longitude" starting at index 2. This could degrade performance. Instead, consider rechunking after loading.
warnings.warn(
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
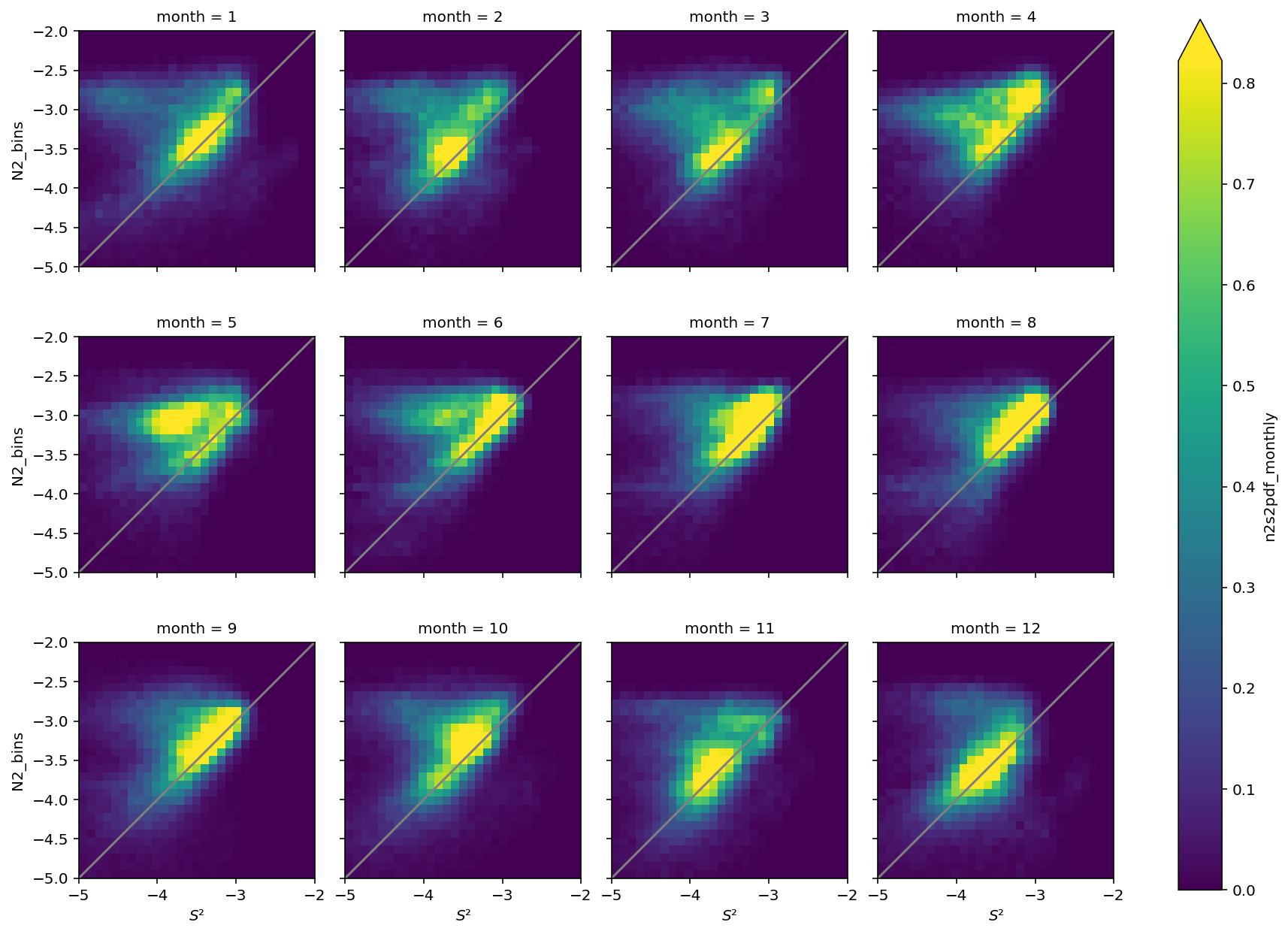
fg = tao_gridded.n2s2pdf_monthly.plot(col="month", col_wrap=4, robust=True)
fg.map(dcpy.plots.line45)
<xarray.plot.facetgrid.FacetGrid at 0x2b2714aeaa40>
Read microstructure#
ls ~/work/datasets/microstructure/osu/
adcp_eq08_30sec.mat T_0_10W_monthly.mat
adcp_eq08.mat T_0_110W_monthly.mat
chipod/ T_0_140W_monthly.mat
chipods_0_10W_hourly.mat T_0_23W_monthly.mat
chipods_0_110W_hourly.mat tao0N140W_make_summary_all_deployments.m
chipods_0_110W.nc th84_timeseries_2009.mat
chipods_0_140W_hourly.mat tiwe.nc
chipods_0_140W.nc tropicheat.nc
chipods_0_23W_hourly.mat tw91_2009.mat
eq08_EUC.mat tw91_sum.mat
eq08_sum_deglitched.mat tw91_velocity.mat
equix/ vel_0_10W_monthly.mat
equix.nc vel_0_110W_monthly.mat
mfiles/ vel_0_140W_monthly.mat
notebooks/ vel_0_23W_monthly.mat
osu_apl_eps.png
micro = mixpods.load_microstructure()
micro
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/xarray/core/computation.py:771: RuntimeWarning: invalid value encountered in log10
result_data = func(*input_data)
<xarray.Dataset>
Dimensions: ()
Data variables:
*empty*datatree.DataTree
- depth: 200
- time: 2624
- zeuc: 80
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- depth(depth)float641.0 2.0 3.0 ... 198.0 199.0 200.0
- positive :
- down
- axis :
- Z
array([ 1., 2., 3., 4., 5., 6., 7., 8., 9., 10., 11., 12., 13., 14., 15., 16., 17., 18., 19., 20., 21., 22., 23., 24., 25., 26., 27., 28., 29., 30., 31., 32., 33., 34., 35., 36., 37., 38., 39., 40., 41., 42., 43., 44., 45., 46., 47., 48., 49., 50., 51., 52., 53., 54., 55., 56., 57., 58., 59., 60., 61., 62., 63., 64., 65., 66., 67., 68., 69., 70., 71., 72., 73., 74., 75., 76., 77., 78., 79., 80., 81., 82., 83., 84., 85., 86., 87., 88., 89., 90., 91., 92., 93., 94., 95., 96., 97., 98., 99., 100., 101., 102., 103., 104., 105., 106., 107., 108., 109., 110., 111., 112., 113., 114., 115., 116., 117., 118., 119., 120., 121., 122., 123., 124., 125., 126., 127., 128., 129., 130., 131., 132., 133., 134., 135., 136., 137., 138., 139., 140., 141., 142., 143., 144., 145., 146., 147., 148., 149., 150., 151., 152., 153., 154., 155., 156., 157., 158., 159., 160., 161., 162., 163., 164., 165., 166., 167., 168., 169., 170., 171., 172., 173., 174., 175., 176., 177., 178., 179., 180., 181., 182., 183., 184., 185., 186., 187., 188., 189., 190., 191., 192., 193., 194., 195., 196., 197., 198., 199., 200.]) - lon(time)float64-139.9 -139.9 ... -139.9 -139.9
- standard_name :
- longitude
- units :
- degrees_east
array([-139.868406, -139.868409, -139.868408, ..., -139.877212, -139.87707 , -139.877121]) - lat(time)float640.06246 0.0622 ... 0.06317 0.06341
- standard_name :
- latitude
- units :
- degrees_north
array([0.062458, 0.062199, 0.062631, ..., 0.063114, 0.063169, 0.063412])
- time(time)datetime64[ns]2008-10-24T20:36:23 ... 2008-11-...
array(['2008-10-24T20:36:23.000000000', '2008-10-24T20:44:18.000000000', '2008-10-24T20:54:17.000000000', ..., '2008-11-08T18:58:49.000000000', '2008-11-08T19:06:14.000000000', '2008-11-08T19:13:47.000000000'], dtype='datetime64[ns]') - zeuc(zeuc)float64-200.0 -195.0 ... 190.0 195.0
- positive :
- up
- long_name :
- $z - z_{EUC}$
- units :
- m
array([-200., -195., -190., -185., -180., -175., -170., -165., -160., -155., -150., -145., -140., -135., -130., -125., -120., -115., -110., -105., -100., -95., -90., -85., -80., -75., -70., -65., -60., -55., -50., -45., -40., -35., -30., -25., -20., -15., -10., -5., 0., 5., 10., 15., 20., 25., 30., 35., 40., 45., 50., 55., 60., 65., 70., 75., 80., 85., 90., 95., 100., 105., 110., 115., 120., 125., 130., 135., 140., 145., 150., 155., 160., 165., 170., 175., 180., 185., 190., 195.]) - enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]])
- pmax(time)float64...
array([205.936515, 199.005106, 202.002421, ..., 202.015744, 221.054025, 203.861253]) - castnumber(time)uint16...
array([ 16, 17, 18, ..., 2666, 2667, 2668], dtype=uint16)
- AX_TILT(depth, time)float64...
[524800 values with dtype=float64]
- AY_TILT(depth, time)float64...
[524800 values with dtype=float64]
- AZ2(depth, time)float64...
[524800 values with dtype=float64]
- C(depth, time)float64...
[524800 values with dtype=float64]
- chi(depth, time)float64...
- long_name :
- $χ$
- units :
- °C²/s
[524800 values with dtype=float64]
- DRHODZ(depth, time)float64...
[524800 values with dtype=float64]
- dTdz(depth, time)float64...
[524800 values with dtype=float64]
- eps(depth, time)float64...
- long_name :
- $ε$
- units :
- W/kg
[524800 values with dtype=float64]
- EPSILON1(depth, time)float64...
[524800 values with dtype=float64]
- EPSILON2(depth, time)float64...
[524800 values with dtype=float64]
- FALLSPD(depth, time)float64...
[524800 values with dtype=float64]
- MHT(depth, time)float64...
[524800 values with dtype=float64]
- N2(depth, time)float64nan nan nan ... 4.678e-05 5.801e-05
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, 8.146444e-06, 5.518243e-05, ..., 1.361518e-05, 1.043197e-05, 1.278190e-05], [ nan, 7.324037e-06, 4.476167e-05, ..., 8.569501e-06, 7.919212e-06, 5.535882e-06], ..., [1.528436e-05, 9.566891e-06, 6.927812e-06, ..., 7.440328e-05, 5.478078e-05, 6.777802e-05], [1.392106e-05, 6.886598e-06, 8.286525e-06, ..., 8.948271e-05, 8.366482e-05, 5.967454e-05], [1.465204e-05, nan, 6.119572e-06, ..., 5.993222e-05, 4.677745e-05, 5.800532e-05]]) - pres(depth)float64...
- standard_name :
- sea_water_pressure
- units :
- dbar
- positive :
- down
array([ nan, nan, nan, nan, nan, nan, nan, nan, nan, 10.059778, 11.065782, 12.071787, 13.077801, 14.083815, 15.089837, 16.09586 , 17.101892, 18.107924, 19.113964, 20.120005, 21.126055, 22.132105, 23.138163, 24.144222, 25.15029 , 26.156358, 27.162434, 28.168511, 29.174597, 30.180683, 31.186777, 32.192872, 33.198976, 34.20508 , 35.211193, 36.217305, 37.223427, 38.229549, 39.23568 , 40.24181 , 41.24795 , 42.25409 , 43.260239, 44.266388, 45.272545, 46.278703, 47.28487 , 48.291037, 49.297213, 50.303388, 51.309573, 52.315758, 53.321952, 54.328146, 55.334348, 56.340551, 57.346763, 58.352975, 59.359196, 60.365416, 61.371646, 62.377876, 63.384115, 64.390354, 65.396602, 66.40285 , 67.409106, 68.415363, 69.421629, 70.427895, 71.43417 , 72.440445, 73.446729, 74.453012, 75.459305, 76.465598, 77.4719 , 78.478202, 79.484513, 80.490824, 81.497144, 82.503464, 83.509792, 84.516121, 85.522459, 86.528797, 87.535144, 88.541491, 89.547847, 90.554203, 91.560568, 92.566933, 93.573307, 94.579681, 95.586064, 96.592447, 97.598839, 98.605231, 99.611632, 100.618033, 101.624443, 102.630853, 103.637272, 104.643691, 105.650119, 106.656547, 107.662984, 108.669421, 109.675867, 110.682313, 111.688768, 112.695223, 113.701687, 114.708151, 115.714624, 116.721098, 117.72758 , 118.734062, 119.740553, 120.747044, 121.753544, 122.760044, 123.766553, 124.773063, 125.779581, 126.786099, 127.792626, 128.799153, 129.805689, 130.812226, 131.818771, 132.825316, 133.83187 , 134.838424, 135.844988, 136.851551, 137.858123, 138.864695, 139.871277, 140.877858, 141.884448, 142.891038, 143.897638, 144.904237, 145.910845, 146.917454, 147.924071, 148.930688, 149.937315, 150.943941, 151.950576, 152.957212, 153.963856, 154.9705 , 155.977154, 156.983807, 157.99047 , 158.997132, 160.003803, 161.010475, 162.017155, 163.023836, 164.030525, 165.037215, 166.043913, 167.050612, 168.057319, 169.064027, 170.070743, 171.07746 , 172.084185, 173.090911, 174.097645, 175.10438 , 176.111124, 177.117867, 178.12462 , 179.131372, 180.138134, 181.144896, 182.151666, 183.158437, 184.165217, 185.171996, 186.178785, 187.185574, 188.192371, 189.199169, 190.205976, 191.212782, 192.219598, 193.226414, 194.233239, 195.240063, 196.246897, 197.253731, 198.260574, 199.267417, 200.274269, 201.28112 ]) - salt(depth, time)float64...
- standard_name :
- sea_water_salinity
- units :
- psu
[524800 values with dtype=float64]
- SCAT(depth, time)float64...
[524800 values with dtype=float64]
- pden(depth, time)float64...
- standard_name :
- sea_water_potential_density
[524800 values with dtype=float64]
- SIGMA_ORDER(depth, time)float64...
[524800 values with dtype=float64]
- T(depth, time)float64...
- standard_name :
- sea_water_temperature
- units :
- celsius
[524800 values with dtype=float64]
- theta(depth, time)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
[524800 values with dtype=float64]
- TP(depth, time)float64...
[524800 values with dtype=float64]
- VARAZ(depth, time)float64...
[524800 values with dtype=float64]
- VARLT(depth, time)float64...
[524800 values with dtype=float64]
- u(depth, time)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_x_velocity
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - v(depth, time)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_y_velocity
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dudz(depth, time)float64...
[524800 values with dtype=float64]
- dvdz(depth, time)float64...
[524800 values with dtype=float64]
- Sh2(depth, time)float64...
[524800 values with dtype=float64]
- sortT(time, depth)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
[524800 values with dtype=float64]
- sortTbyT(time, depth)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
[524800 values with dtype=float64]
- Jq(depth, time)float64...
- long_name :
- $J_q^ε$
- units :
- W/m²
[524800 values with dtype=float64]
- dJdz(depth, time)float64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
[524800 values with dtype=float64]
- dTdt(depth, time)float64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
[524800 values with dtype=float64]
- eucmax(time)float64nan nan nan nan ... nan nan nan nan
- positive :
- down
- long_name :
- Depth of EUC max
- units :
- m
array([nan, nan, nan, ..., nan, nan, nan])
- mld(time)float64...
- long_name :
- MLD
- units :
- m
- description :
- Interpolate density to 1m grid. Search for min depth where |drho| > 0.005 and N2 > 1e-08
array([ 2., 6., 3., ..., 13., 9., 12.])
- Jq_euc(time, zeuc)float64...
- long_name :
- $J_q^ε$
- units :
- W/m²
[209920 values with dtype=float64]
- dJdz_euc(time, zeuc)float64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
[209920 values with dtype=float64]
- dTdt_euc(time, zeuc)float64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
[209920 values with dtype=float64]
- u_euc(time, zeuc)float64...
[209920 values with dtype=float64]
- depth_euc(time, zeuc)float64...
- positive :
- down
- axis :
- Z
[209920 values with dtype=float64]
- count_Jq_euc(time, zeuc)int64...
- long_name :
- $J_q^ε$
- units :
- W/m²
[209920 values with dtype=int64]
- count_dJdz_euc(time, zeuc)int64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
[209920 values with dtype=int64]
- count_dTdt_euc(time, zeuc)int64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
[209920 values with dtype=int64]
- count_u_euc(time, zeuc)int64...
[209920 values with dtype=int64]
- count_depth_euc(time, zeuc)int64...
- positive :
- down
- axis :
- Z
[209920 values with dtype=int64]
- S2(depth, time)float64nan nan nan nan ... nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - shred2(depth, time)float64nan nan nan nan ... nan nan nan nan
- long_name :
- $Sh_{red}^2$
- units :
- $s^{-2}$
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - Ri(depth, time)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Ri
- units :
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - n2s2pdf(enso_transition_phase, N2_bins, S2_bins)float640.03719 0.04226 0.04184 ... 0.0 0.0
- long_name :
- $P(S^2, 4N^2)$
array([[[3.71928280e-02, 4.22645773e-02, 4.18419315e-02, 4.56457434e-02, 5.19854300e-02, 3.71928280e-02, 3.63475364e-02, 4.14192857e-02, 3.38116618e-02, 2.57813921e-02, 2.28228717e-02, 1.14114359e-02, 6.33968659e-03, 7.18497813e-03, 3.80381195e-03, 1.26793732e-03, 4.22645773e-04, 2.53587464e-03, 4.22645773e-04, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [4.90269096e-02, 5.53665962e-02, 4.86042638e-02, 6.04383455e-02, 6.38195117e-02, 7.18497813e-02, 5.79024708e-02, 6.84686152e-02, 5.70571793e-02, 4.22645773e-02, 2.78946210e-02, 1.81737682e-02, 9.72085277e-03, 4.22645773e-03, 4.64910350e-03, 2.53587464e-03, 8.45291545e-04, 0.00000000e+00, 1.26793732e-03, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [6.63553863e-02, 6.72006778e-02, 6.50874490e-02, 7.48083017e-02, 7.98800510e-02, 9.46726531e-02, 9.17141327e-02, 9.29820700e-02, 7.69215306e-02, 5.62118878e-02, 5.40986589e-02, 3.38116618e-02, 2.15549344e-02, 1.31020190e-02, 5.91704082e-03, 2.95852041e-03, ... 5.49439504e-03, 2.95852041e-03, 9.29820700e-03, 1.90190598e-02, 2.24002259e-02, 2.19775802e-02, 2.15549344e-02, 8.87556122e-03, 2.53587464e-03, 4.22645773e-04, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 8.45291545e-04, 4.22645773e-04, 8.45291545e-04, 1.69058309e-03, 2.95852041e-03, 6.76233236e-03, 6.76233236e-03, 1.35246647e-02, 1.73284767e-02, 1.01434985e-02, 3.80381195e-03, 0.00000000e+00, 4.22645773e-04, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 4.22645773e-04, 0.00000000e+00, 0.00000000e+00, 1.26793732e-03, 4.22645773e-04, 3.80381195e-03, 5.07174927e-03, 7.18497813e-03, 5.91704082e-03, 1.26793732e-03, 8.45291545e-04, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]])
- starttime :
- ['Time:20:34:29 298 ' 'Time:20:42:18 298 ' 'Time:20:52:14 298 ' ... 'Time:18:56:50 313 ' 'Time:19:04:01 313 ' 'Time:19:11:46 313 ']
- endtime :
- ['Time:20:38:29 298 ' 'Time:20:46:29 298 ' 'Time:20:56:29 298 ' ... 'Time:19:01:00 313 ' 'Time:19:08:40 313 ' 'Time:19:16:00 313 ']
- time: 287
- depth: 60
- zeuc: 80
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- time(time)datetime64[ns]1984-11-19T20:30:02 ... 1984-12-...
array(['1984-11-19T20:30:02.000000000', '1984-11-19T21:29:57.000000000', '1984-11-19T22:30:00.000000000', ..., '1984-12-01T16:25:49.000000000', '1984-12-01T17:25:52.000000000', '1984-12-01T18:25:46.000000000'], dtype='datetime64[ns]') - lat(time)float64-0.0355 -0.012 ... 0.0028 -0.0127
- standard_name :
- latitude
- units :
- degrees_north
array([-0.0355, -0.012 , -0.0043, ..., -0.012 , 0.0028, -0.0127])
- lon(time)float64140.0 140.0 140.0 ... 140.0 140.0
- standard_name :
- longitude
- units :
- degrees_east
array([140., 140., 140., ..., 140., 140., 140.])
- depth(depth)float643.1 7.1 11.1 ... 231.1 235.1 239.1
- positive :
- down
- axis :
- Z
array([ 3.1, 7.1, 11.1, 15.1, 19.1, 23.1, 27.1, 31.1, 35.1, 39.1, 43.1, 47.1, 51.1, 55.1, 59.1, 63.1, 67.1, 71.1, 75.1, 79.1, 83.1, 87.1, 91.1, 95.1, 99.1, 103.1, 107.1, 111.1, 115.1, 119.1, 123.1, 127.1, 131.1, 135.1, 139.1, 143.1, 147.1, 151.1, 155.1, 159.1, 163.1, 167.1, 171.1, 175.1, 179.1, 183.1, 187.1, 191.1, 195.1, 199.1, 203.1, 207.1, 211.1, 215.1, 219.1, 223.1, 227.1, 231.1, 235.1, 239.1]) - zeuc(zeuc)float64-200.0 -195.0 ... 190.0 195.0
- positive :
- up
- long_name :
- $z - z_{EUC}$
- units :
- m
array([-200., -195., -190., -185., -180., -175., -170., -165., -160., -155., -150., -145., -140., -135., -130., -125., -120., -115., -110., -105., -100., -95., -90., -85., -80., -75., -70., -65., -60., -55., -50., -45., -40., -35., -30., -25., -20., -15., -10., -5., 0., 5., 10., 15., 20., 25., 30., 35., 40., 45., 50., 55., 60., 65., 70., 75., 80., 85., 90., 95., 100., 105., 110., 115., 120., 125., 130., 135., 140., 145., 150., 155., 160., 165., 170., 175., 180., 185., 190., 195.]) - enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]])
- wspeed(time)float64...
array([7.54 , 6.93 , 6.71 , ..., 8.635, 8.625, 8.445])
- T(depth, time)float64...
- standard_name :
- sea_water_temperature
- units :
- celsius
array([[24.1577 , 24.1824 , 24.243 , ..., 24.9522 , 24.9345 , 24.898899], [24.103201, 24.0872 , 24.0912 , ..., 24.9529 , 24.9347 , 24.8894 ], [23.989901, 24.0112 , 24.0287 , ..., 24.953899, 24.931601, 24.8911 ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - salt(depth, time)float64...
- standard_name :
- sea_water_salinity
- units :
- psu
array([[34.8148 , 34.817001, 34.813599, ..., 35.1404 , 35.1441 , 35.1394 ], [34.815498, 34.816002, 34.8125 , ..., 35.140202, 35.144501, 35.138199], [34.818901, 34.818001, 34.814499, ..., 35.140301, 35.1446 , 35.138901], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - pden(depth, time)float64...
- standard_name :
- sea_water_potential_density
array([[1023.4585 , 1023.452801, 1023.4321 , ..., 1023.4667 , 1023.4748 , 1023.481899], [1023.4751 , 1023.480301, 1023.4764 , ..., 1023.466299, 1023.475 , 1023.4841 ], [1023.511299, 1023.504299, 1023.496599, ..., 1023.466 , 1023.476101, 1023.4841 ], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - u(depth, time)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_x_velocity
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - v(depth, time)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_y_velocity
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - eps(depth, time)float64...
- long_name :
- $ε$
- units :
- W/kg
array([[ nan, nan, nan, ..., 6.3798e-05, 7.6077e-05, nan], [ nan, nan, nan, ..., 1.1682e-06, 2.7831e-06, nan], [ nan, nan, nan, ..., 7.5588e-08, 4.2374e-07, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - dTdz(depth, time)float64...
array([[-1.362475e-02, -2.380000e-02, -3.795000e-02, ..., 1.750000e-04, 5.000000e-05, -2.374750e-03], [-2.097487e-02, -2.140000e-02, -2.678750e-02, ..., 2.123750e-04, -3.623750e-04, -9.748750e-04], [-2.447512e-02, -1.880000e-02, -1.828737e-02, ..., -2.375000e-04, -1.212625e-03, -1.150125e-03], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - dsigdz(depth, time)float64...
array([[ 4.150000e-03, 6.875000e-03, 1.107500e-02, ..., -1.002500e-04, 5.000000e-05, 5.502500e-04], [ 6.599875e-03, 6.437250e-03, 8.062375e-03, ..., -8.750000e-05, 1.626250e-04, 2.751250e-04], [ 8.275000e-03, 6.374875e-03, 6.187500e-03, ..., 7.512500e-05, 3.875000e-04, 2.625000e-04], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - N2(depth, time)float643.976e-05 6.586e-05 ... nan nan
array([[3.975732e-05, 6.586304e-05, 1.060994e-04, ..., nan, 4.790039e-07, 5.271438e-06], [6.322732e-05, 6.166936e-05, 7.723818e-05, ..., nan, 1.557960e-06, 2.635719e-06], [7.927515e-05, 6.107180e-05, 5.927673e-05, ..., 7.197034e-07, 3.712280e-06, 2.514771e-06], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - dudz(depth, time)float64...
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dvdz(depth, time)float64...
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - pres(depth)float64...
- standard_name :
- sea_water_pressure
- units :
- dbar
array([ 3.118483, 7.142396, 11.166381, 15.190437, 19.214566, 23.238767, 27.26304 , 31.287385, 35.311802, 39.33629 , 43.360851, 47.385484, 51.410189, 55.434966, 59.459815, 63.484737, 67.50973 , 71.534795, 75.559932, 79.585142, 83.610423, 87.635777, 91.661202, 95.6867 , 99.71227 , 103.737911, 107.763625, 111.789411, 115.81527 , 119.8412 , 123.867202, 127.893276, 131.919423, 135.945642, 139.971932, 143.998295, 148.02473 , 152.051238, 156.077817, 160.104468, 164.131192, 168.157988, 172.184856, 176.211796, 180.238808, 184.265892, 188.293049, 192.320278, 196.347578, 200.374951, 204.402397, 208.429914, 212.457504, 216.485166, 220.5129 , 224.540706, 228.568584, 232.596535, 236.624558, 240.652653]) - theta(depth, time)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
array([[24.157042, 24.181741, 24.24234 , ..., 24.951525, 24.933826, 24.898225], [24.101696, 24.085696, 24.089696, ..., 24.951354, 24.933155, 24.887857], [23.987556, 24.008854, 24.026352, ..., 24.951482, 24.929186, 24.888688], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - sortT(time, depth)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
array([[24.157042, 24.101696, 23.987556, ..., nan, nan, nan], [24.181741, 24.085696, 24.008854, ..., nan, nan, nan], [24.24234 , 24.089696, 24.026352, ..., nan, nan, nan], ..., [24.951482, 24.951354, 24.951525, ..., nan, nan, nan], [24.933826, 24.933155, 24.929186, ..., nan, nan, nan], [24.898225, 24.887857, 24.888688, ..., nan, nan, nan]]) - sortTbyT(time, depth)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
array([[24.157042, 24.101696, 23.987556, ..., nan, nan, nan], [24.181741, 24.085696, 24.008854, ..., nan, nan, nan], [24.24234 , 24.089696, 24.026352, ..., nan, nan, nan], ..., [24.951525, 24.951482, 24.951354, ..., nan, nan, nan], [24.933826, 24.933155, 24.929186, ..., nan, nan, nan], [24.898225, 24.888688, 24.887857, ..., nan, nan, nan]]) - Jq(depth, time)float64...
- long_name :
- $J_q^ε$
- units :
- W/m²
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, -530.816644, nan], [ nan, nan, nan, ..., nan, -113.50084 , nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - dJdz(depth, time)float64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, -9.638447, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - dTdt(depth, time)float64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, 6.093379, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]) - eucmax(time)float64119.1 119.1 119.1 ... 107.1 107.1
- positive :
- down
- long_name :
- Depth of EUC max
- units :
- m
array([119.1, 119.1, 119.1, nan, 115.1, 119.1, 119.1, 119.1, 119.1, 115.1, 123.1, 123.1, 123.1, 123.1, 127.1, 127.1, 127.1, 123.1, 123.1, 127.1, 135.1, 139.1, 135.1, 131.1, 131.1, nan, nan, nan, nan, 139.1, 139.1, nan, 147.1, 143.1, 143.1, 147.1, 151.1, 151.1, 151.1, 147.1, 147.1, 147.1, nan, 147.1, 151.1, 151.1, 151.1, 147.1, nan, nan, nan, nan, nan, 143.1, 151.1, 155.1, 151.1, 151.1, 151.1, 151.1, 147.1, 147.1, 143.1, 143.1, 139.1, 139.1, 143.1, 139.1, 139.1, 139.1, 135.1, 135.1, 131.1, 127.1, 123.1, 123.1, 123.1, 123.1, 123.1, 123.1, 123.1, 123.1, 119.1, 119.1, 119.1, 123.1, 119.1, 115.1, 111.1, 111.1, 115.1, 115.1, 119.1, 119.1, 119.1, 119.1, 115.1, nan, nan, 107.1, 107.1, 103.1, 107.1, 111.1, 115.1, nan, 119.1, nan, 115.1, 111.1, 115.1, 123.1, 123.1, 115.1, 111.1, 111.1, 111.1, 111.1, 115.1, 115.1, nan, nan, nan, 123.1, 119.1, 123.1, nan, nan, nan, nan, 115.1, 115.1, 119.1, 131.1, 123.1, 119.1, 111.1, 111.1, 111.1, 107.1, 103.1, 103.1, 103.1, 103.1, nan, 107.1, 111.1, 115.1, 115.1, 111.1, 107.1, 107.1, 107.1, 107.1, 111.1, 119.1, 115.1, 119.1, 115.1, 111.1, 111.1, 111.1, 107.1, 107.1, 107.1, 107.1, 111.1, 115.1, 119.1, 119.1, 115.1, 115.1, 115.1, 115.1, 115.1, 111.1, 107.1, 107.1, 107.1, 115.1, 115.1, 119.1, 115.1, 115.1, 115.1, 111.1, 107.1, 107.1, 111.1, 111.1, 111.1, 115.1, 119.1, 123.1, 119.1, 115.1, 111.1, 107.1, 107.1, 103.1, 107.1, 107.1, 103.1, 103.1, 107.1, 111.1, 111.1, 111.1, 115.1, 111.1, 111.1, 111.1, 111.1, 111.1, 111.1, nan, 115.1, 119.1, 123.1, 123.1, 119.1, 119.1, 115.1, 119.1, 123.1, 119.1, 119.1, 119.1, 123.1, 127.1, 131.1, 131.1, 135.1, 131.1, 127.1, 127.1, 131.1, 131.1, nan, 123.1, 123.1, 123.1, 123.1, 123.1, 123.1, 119.1, 123.1, 123.1, 127.1, 123.1, 119.1, 115.1, 115.1, 115.1, 123.1, 123.1, 119.1, 119.1, 123.1, 119.1, 119.1, 123.1, nan, 119.1, 115.1, 115.1, 119.1, 119.1, 123.1, 123.1, 119.1, 119.1, 119.1, 119.1, 119.1, 115.1, 115.1, 115.1, 111.1, 111.1, 107.1, 111.1, 111.1, 111.1, 111.1, 107.1, 107.1]) - mld(time)float64...
- long_name :
- MLD
- units :
- m
- description :
- Interpolate density to 1m grid. Search for min depth where |drho| > 0.005 and N2 > 1e-08
array([ 7.1, 7.1, 7.1, ..., 23.1, 19.1, 19.1])
- gamma_n(time, depth)float64...
- standard_name :
- neutral_density
- units :
- kg/m3
- long_name :
- $γ_n$
array([[23.464375, 23.481443, 23.518007, ..., nan, nan, nan], [23.458675, 23.48659 , 23.511002, ..., nan, nan, nan], [23.438003, 23.482748, 23.503151, ..., nan, nan, nan], ..., [23.471187, 23.471109, 23.471165, ..., nan, nan, nan], [23.47938 , 23.479907, 23.48121 , ..., nan, nan, nan], [23.486665, 23.488932, 23.48923 , ..., nan, nan, nan]]) - Jq_euc(time, zeuc)float64...
- long_name :
- $J_q^ε$
- units :
- W/m²
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dJdz_euc(time, zeuc)float64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - dTdt_euc(time, zeuc)float64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - u_euc(time, zeuc)float64...
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - count_Jq_euc(time, zeuc)int64...
- long_name :
- $J_q^ε$
- units :
- W/m²
array([[0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], ..., [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0]]) - count_dJdz_euc(time, zeuc)int64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
array([[0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], ..., [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0]]) - count_dTdt_euc(time, zeuc)int64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
array([[0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], ..., [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0]]) - count_u_euc(time, zeuc)int64...
array([[0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], ..., [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0], [0, 0, 0, ..., 0, 0, 0]]) - S2(depth, time)float64nan nan nan nan ... nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - shred2(depth, time)float64nan nan nan nan ... nan nan nan nan
- long_name :
- $Sh_{red}^2$
- units :
- $s^{-2}$
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - Ri(depth, time)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Ri
- units :
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - n2s2pdf(enso_transition_phase, N2_bins, S2_bins)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
- long_name :
- $P(S^2, 4N^2)$
array([[[0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ], [0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ], [0. , 0.01625121, 0.01625121, 0.01625121, 0. , 0.01625121, 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ], [0. , 0. , 0. , 0. , 0.01625121, 0. , 0. , 0. , 0. , 0. , ... 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ], [0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ], [0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ], [0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. , 0. ]]])
- readme :
- ['this file is compiled from a binary ' 'Fortran file (converted to ascii beforehand):' 'th84ts.m4 --> th84ts.txt ']
- name :
- Tropic Heat
- depth: 250
- time: 3776
- zeuc: 80
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- depth(depth)float641.0 2.0 3.0 ... 248.0 249.0 250.0
- axis :
- Z
- positive :
- down
array([ 1., 2., 3., ..., 248., 249., 250.])
- time(time)datetime64[ns]1991-11-04T18:43:50 ... 1991-11-...
- axis :
- T
- standard_name :
- time
array(['1991-11-04T18:43:50.000000000', '1991-11-04T18:46:51.000000000', '1991-11-04T18:53:21.000000000', ..., '1991-11-24T22:51:04.000000000', '1991-11-24T22:57:34.000000000', '1991-11-24T23:04:04.000000000'], dtype='datetime64[ns]') - latitude()int640
- units :
- degrees_north
- standard_name :
- latitude
array(0)
- longitude()int64-140
- units :
- degrees_east
- standard_name :
- longitude
array(-140)
- zeuc(zeuc)float64-200.0 -195.0 ... 190.0 195.0
- positive :
- up
- long_name :
- $z - z_{EUC}$
- units :
- m
array([-200., -195., -190., -185., -180., -175., -170., -165., -160., -155., -150., -145., -140., -135., -130., -125., -120., -115., -110., -105., -100., -95., -90., -85., -80., -75., -70., -65., -60., -55., -50., -45., -40., -35., -30., -25., -20., -15., -10., -5., 0., 5., 10., 15., 20., 25., 30., 35., 40., 45., 50., 55., 60., 65., 70., 75., 80., 85., 90., 95., 100., 105., 110., 115., 120., 125., 130., 135., 140., 145., 150., 155., 160., 165., 170., 175., 180., 185., 190., 195.]) - enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]])
- chi(depth, time)float64...
- long_name :
- $χ$
- units :
- °C²/s
[944000 values with dtype=float64]
- pres(depth)float64...
- standard_name :
- sea_water_pressure
- units :
- dbar
array([ nan, nan, nan, ..., 249.61546 , 250.622533, 251.629606]) - salt(depth, time)float64...
- standard_name :
- sea_water_salinity
- units :
- psu
[944000 values with dtype=float64]
- pden(depth, time)float64...
- standard_name :
- sea_water_potential_density
[944000 values with dtype=float64]
- T(depth, time)float64...
- standard_name :
- sea_water_temperature
- units :
- celsius
[944000 values with dtype=float64]
- theta(depth, time)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
[944000 values with dtype=float64]
- eps(depth, time)float64...
- long_name :
- $ε$
- units :
- W/kg
[944000 values with dtype=float64]
- EPSILON_clean(depth, time)float64...
[944000 values with dtype=float64]
- u(depth, time)float64nan nan nan ... 0.03472 0.03476
- standard_name :
- sea_water_x_velocity
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., 0.047633, 0.046282, 0.045933], [ nan, nan, nan, ..., 0.041579, 0.0405 , 0.040347], [ nan, nan, nan, ..., 0.035526, 0.034717, 0.034762]]) - v(depth, time)float64nan nan nan ... 0.02746 0.02942
- standard_name :
- sea_water_y_velocity
array([[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., 0.023839, 0.026215, 0.028178], [ nan, nan, nan, ..., 0.024451, 0.026839, 0.028796], [ nan, nan, nan, ..., 0.025063, 0.027463, 0.029415]]) - dTdz(depth, time)float64...
- units :
- celsius
[944000 values with dtype=float64]
- N2(depth, time)float64nan nan nan nan ... nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - sortT(time, depth)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
[944000 values with dtype=float64]
- sortTbyT(time, depth)float64...
- standard_name :
- sea_water_potential_temperature
- units :
- celsius
[944000 values with dtype=float64]
- Jq(depth, time)float64...
- long_name :
- $J_q^ε$
- units :
- W/m²
[944000 values with dtype=float64]
- dJdz(depth, time)float64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
[944000 values with dtype=float64]
- dTdt(depth, time)float64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
[944000 values with dtype=float64]
- eucmax(time)float64nan nan nan ... 108.0 108.0 108.0
- positive :
- down
- long_name :
- Depth of EUC max
- units :
- m
array([ nan, nan, nan, ..., 108., 108., 108.])
- mld(time)float64...
- long_name :
- MLD
- units :
- m
- description :
- Interpolate density to 1m grid. Search for min depth where |drho| > 0.005 and N2 > 1e-08
array([nan, nan, nan, ..., 13., 10., 14.])
- gamma_n(time, depth)float64...
- standard_name :
- neutral_density
- units :
- kg/m3
- long_name :
- $γ_n$
[944000 values with dtype=float64]
- Jq_euc(time, zeuc)float64...
- long_name :
- $J_q^ε$
- units :
- W/m²
[302080 values with dtype=float64]
- dJdz_euc(time, zeuc)float64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
[302080 values with dtype=float64]
- dTdt_euc(time, zeuc)float64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
[302080 values with dtype=float64]
- u_euc(time, zeuc)float64...
[302080 values with dtype=float64]
- count_Jq_euc(time, zeuc)int64...
- long_name :
- $J_q^ε$
- units :
- W/m²
[302080 values with dtype=int64]
- count_dJdz_euc(time, zeuc)int64...
- long_name :
- $∂J_q^ε/∂z$
- units :
- W/kg/m
[302080 values with dtype=int64]
- count_dTdt_euc(time, zeuc)int64...
- long_name :
- $∂T/∂t = -1/(ρ_0c_p) ∂J_q^ε/∂z$
- units :
- °C/month
[302080 values with dtype=int64]
- count_u_euc(time, zeuc)int64...
[302080 values with dtype=int64]
- S2(depth, time)float64nan nan nan nan ... nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - shred2(depth, time)float64nan nan nan nan ... nan nan nan nan
- long_name :
- $Sh_{red}^2$
- units :
- $s^{-2}$
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - Ri(depth, time)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Ri
- units :
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - n2s2pdf(enso_transition_phase, N2_bins, S2_bins)float640.01279 0.01681 0.01389 ... 0.0 0.0
- long_name :
- $P(S^2, 4N^2)$
array([[[1.27924977e-02, 1.68129970e-02, 1.38889975e-02, 2.01024964e-02, 2.04679964e-02, 1.75439969e-02, 2.63159953e-02, 2.81434950e-02, 4.53219920e-02, 4.23979925e-02, 5.22664907e-02, 5.44594904e-02, 3.03364946e-02, 2.99709947e-02, 1.90059966e-02, 9.13749838e-03, 2.55849955e-03, 1.09649981e-03, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.71784970e-02, 1.42544975e-02, 2.11989962e-02, 2.22954960e-02, 1.90059966e-02, 2.70469952e-02, 2.55849955e-02, 3.72809934e-02, 4.64184918e-02, 6.76174880e-02, 7.34654870e-02, 6.46934885e-02, 5.33629905e-02, 4.82459915e-02, 2.55849955e-02, 1.16959979e-02, 1.82749968e-03, 1.09649981e-03, 0.00000000e+00, 3.65499935e-04, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [2.08334963e-02, 2.15644962e-02, 1.86404967e-02, 2.63159953e-02, 2.33919959e-02, 3.69154935e-02, 3.91084931e-02, 5.11699909e-02, 7.05414875e-02, 7.52929867e-02, 7.71204863e-02, 9.02784840e-02, 6.32314888e-02, 6.21349890e-02, 4.09359927e-02, 2.01024964e-02, ... 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [7.30999870e-04, 7.30999870e-04, 3.65499935e-04, 7.30999870e-04, 7.30999870e-04, 4.02049929e-03, 0.00000000e+00, 1.82749968e-03, 5.48249903e-03, 6.21349890e-03, 4.75149916e-03, 8.40649851e-03, 1.02339982e-02, 6.57899883e-03, 2.55849955e-03, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 7.30999870e-04, 7.30999870e-04, 2.19299961e-03, 1.09649981e-03, 2.19299961e-03, 3.65499935e-04, 7.30999870e-04, 1.09649981e-03, 7.30999870e-04, 7.30999870e-04, 4.02049929e-03, 8.04099858e-03, 7.67549864e-03, 5.84799896e-03, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]])
- starttime :
- ['' '' '' ... 'Time:22:51:04 328 ' 'Time:22:57:35 328 ' 'Time:23:04:04 328 ']
- endtime :
- ['' '' '' ... 'Time:22:54:39 328 ' 'Time:23:01:10 328 ' 'Time:23:07:49 328 ']
- readme :
- ['EPSILON_clean cleaned using tw91_eps_chi_sum1.mat ' '(all that is marked NaN or missed in tw91_eps_chi_sum1.mat ' 'is marked NaN in that field too) plus bad_drops.40, ' 'which contained contaminated casts, is used to mark bad EPSILON']
- name :
- TIWE
<xarray.Dataset> Dimensions: (depth: 200, time: 2624, zeuc: 80, enso_transition_phase: 1, N2_bins: 29, S2_bins: 29) Coordinates: * depth (depth) float64 1.0 2.0 3.0 4.0 ... 198.0 199.0 200.0 lon (time) float64 -139.9 -139.9 -139.9 ... -139.9 -139.9 lat (time) float64 0.06246 0.0622 ... 0.06317 0.06341 * time (time) datetime64[ns] 2008-10-24T20:36:23 ... 2008... * zeuc (zeuc) float64 -200.0 -195.0 -190.0 ... 190.0 195.0 * enso_transition_phase (enso_transition_phase) object 'none' * N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-... * S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-... bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107 Data variables: (12/51) pmax (time) float64 205.9 199.0 202.0 ... 221.1 203.9 castnumber (time) uint16 16 17 18 19 20 ... 2665 2666 2667 2668 AX_TILT (depth, time) float64 ... AY_TILT (depth, time) float64 ... AZ2 (depth, time) float64 ... C (depth, time) float64 ... ... ... count_u_euc (time, zeuc) int64 ... count_depth_euc (time, zeuc) int64 ... S2 (depth, time) float64 nan nan nan nan ... nan nan nan shred2 (depth, time) float64 nan nan nan nan ... nan nan nan Ri (depth, time) float64 nan nan nan nan ... nan nan nan n2s2pdf (enso_transition_phase, N2_bins, S2_bins) float64 ... Attributes: starttime: ['Time:20:34:29 298 ' 'Time:20:42:18 298 ' 'Time:20:52:14... endtime: ['Time:20:38:29 298 ' 'Time:20:46:29 298 ' 'Time:20:56:29...equix<xarray.Dataset> Dimensions: (time: 287, depth: 60, zeuc: 80, enso_transition_phase: 1, N2_bins: 29, S2_bins: 29) Coordinates: * time (time) datetime64[ns] 1984-11-19T20:30:02 ... 1984... lat (time) float64 -0.0355 -0.012 ... 0.0028 -0.0127 lon (time) float64 140.0 140.0 140.0 ... 140.0 140.0 * depth (depth) float64 3.1 7.1 11.1 ... 231.1 235.1 239.1 * zeuc (zeuc) float64 -200.0 -195.0 -190.0 ... 190.0 195.0 * enso_transition_phase (enso_transition_phase) object 'none' * N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-... * S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-... bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107 Data variables: (12/34) wspeed (time) float64 7.54 6.93 6.71 ... 8.635 8.625 8.445 T (depth, time) float64 24.16 24.18 24.24 ... nan nan salt (depth, time) float64 34.81 34.82 34.81 ... nan nan pden (depth, time) float64 1.023e+03 1.023e+03 ... nan nan u (depth, time) float64 nan nan nan nan ... nan nan nan v (depth, time) float64 nan nan nan nan ... nan nan nan ... ... count_dTdt_euc (time, zeuc) int64 0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 count_u_euc (time, zeuc) int64 0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 S2 (depth, time) float64 nan nan nan nan ... nan nan nan shred2 (depth, time) float64 nan nan nan nan ... nan nan nan Ri (depth, time) float64 nan nan nan nan ... nan nan nan n2s2pdf (enso_transition_phase, N2_bins, S2_bins) float64 ... Attributes: readme: ['this file is compiled from a binary '\n 'Fortran fil... name: Tropic Heattropicheat<xarray.Dataset> Dimensions: (depth: 250, time: 3776, zeuc: 80, enso_transition_phase: 1, N2_bins: 29, S2_bins: 29) Coordinates: * depth (depth) float64 1.0 2.0 3.0 4.0 ... 248.0 249.0 250.0 * time (time) datetime64[ns] 1991-11-04T18:43:50 ... 1991... latitude int64 0 longitude int64 -140 * zeuc (zeuc) float64 -200.0 -195.0 -190.0 ... 190.0 195.0 * enso_transition_phase (enso_transition_phase) object 'none' * N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-... * S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-... bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107 Data variables: (12/32) chi (depth, time) float64 ... pres (depth) float64 nan nan nan nan ... 249.6 250.6 251.6 salt (depth, time) float64 ... pden (depth, time) float64 ... T (depth, time) float64 ... theta (depth, time) float64 ... ... ... count_dTdt_euc (time, zeuc) int64 ... count_u_euc (time, zeuc) int64 ... S2 (depth, time) float64 nan nan nan nan ... nan nan nan shred2 (depth, time) float64 nan nan nan nan ... nan nan nan Ri (depth, time) float64 nan nan nan nan ... nan nan nan n2s2pdf (enso_transition_phase, N2_bins, S2_bins) float64 ... Attributes: starttime: ['' '' '' ... 'Time:22:51:04 328 ' 'Time:22:57:35 328 '\n... endtime: ['' '' '' ... 'Time:22:54:39 328 ' 'Time:23:01:10 328 '\n... readme: ['EPSILON_clean cleaned using tw91_eps_chi_sum1.mat ... name: TIWEtiwe
Stability Diagram#
Contours enclose 50%, 75% of the data
Colors are for LES
White contours are 50%, 75% contours for TAO 0, 140
thnk about LES resampling?
Domain averages vs moorings#
Averaging over the domain (red) tightens the scatter plot along the 1:1 line
f, ax = plt.subplots(1, 4, sharex=True, sharey=True, constrained_layout=True)
for lat, axx in zip(moorings.ds["latitude"].data, ax):
mixpods.plot_n2s2pdf(
moorings.ds.n2s2pdf.sel(enso_transition_phase="none", latitude=lat),
ax=axx,
pcolor=False,
colors="k",
)
mixpods.plot_n2s2pdf(
avgs.ds.n2s2pdf.sel(enso_transition_phase="none", latitude=lat),
ax=axx,
pcolor=False,
colors="r",
)
axx.set_title(f"latitude={lat}")
dcpy.plots.clean_axes(ax)
f.set_size_inches((8, 6))
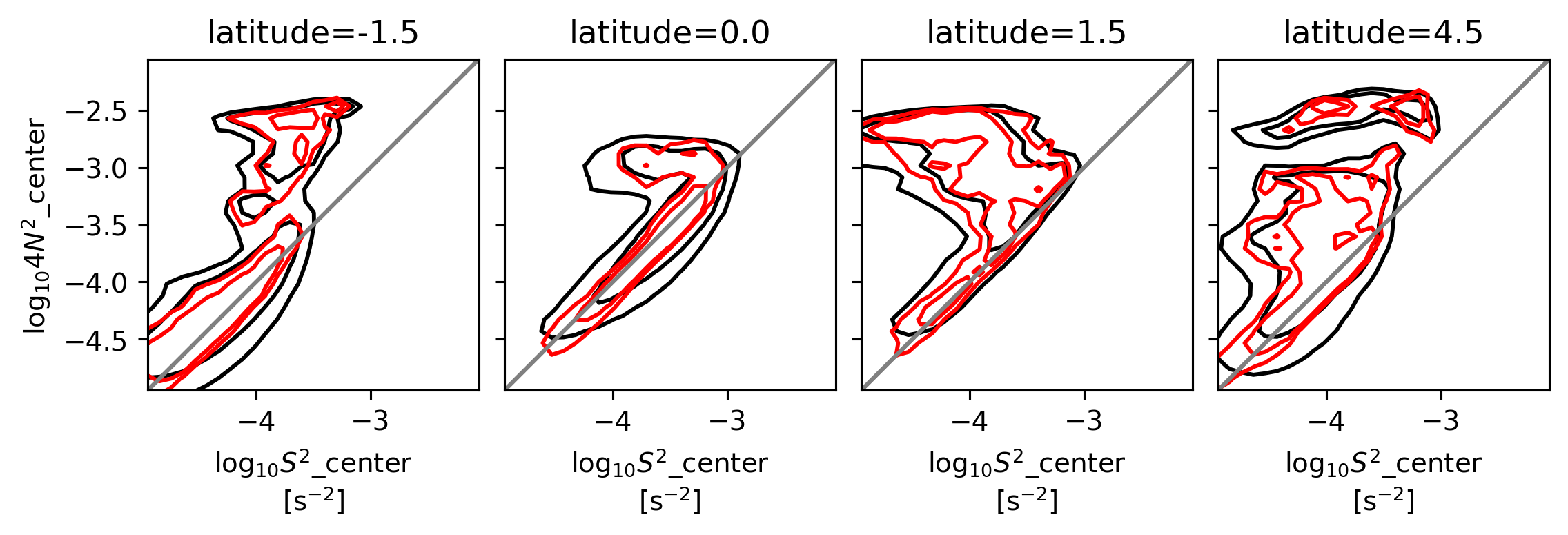
vs TAO#
f, ax = plt.subplots(1, 4, sharex=True, sharey=True, constrained_layout=True)
for lat, axx in zip(moorings.ds["latitude"].data, ax):
mixpods.plot_n2s2pdf(
moorings.ds.n2s2pdf.sel(latitude=lat),
ax=axx,
vmin=0,
vmax=0.8,
add_colorbar=False,
cmap="viridis",
)
mixpods.plot_n2s2pdf(
tao_gridded.n2s2pdf.sel(enso_transition_phase="none"),
ax=axx,
pcolor=False,
colors="w",
)
axx.set_title(f"latitude={lat}")
dcpy.plots.clean_axes(ax)
f.set_size_inches((8, 6))
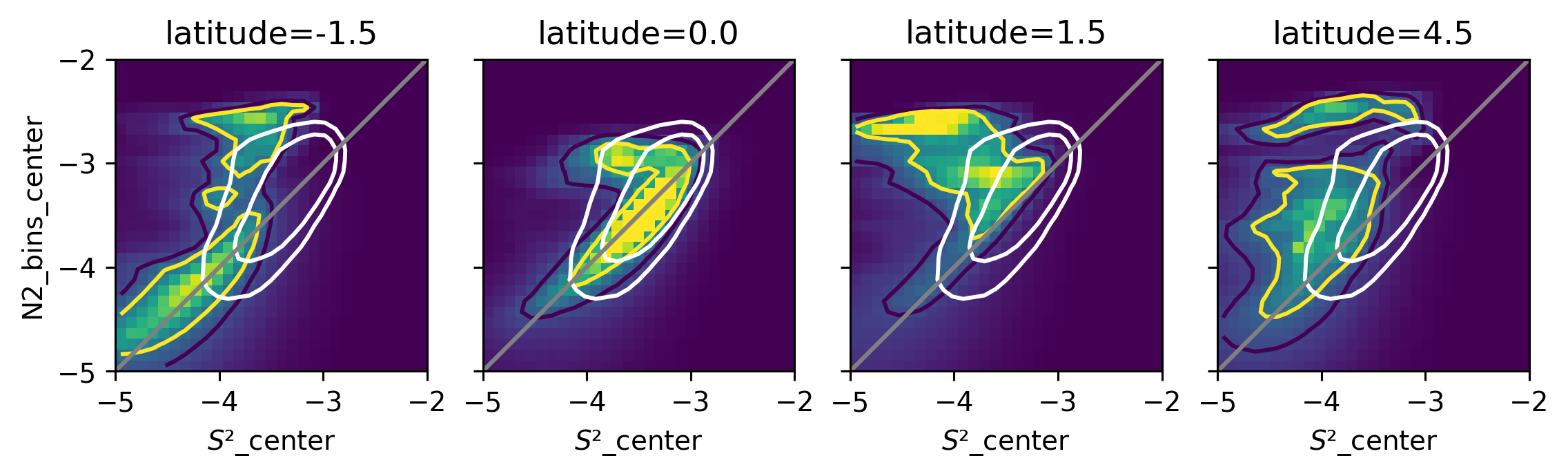
vs TAO in October#
Comparing October contours only in TAO makes it look better
f, ax = plt.subplots(1, 4, sharex=True, sharey=True, constrained_layout=True)
for lat, axx in zip(moorings.ds.latitude.data, ax):
mixpods.plot_n2s2pdf(
moorings.ds.n2s2pdf.sel(latitude=lat),
ax=axx,
vmin=0,
vmax=0.8,
add_colorbar=False,
cmap="viridis",
)
mixpods.plot_n2s2pdf(
tao_gridded.n2s2pdf_monthly.sel(month=10), ax=axx, pcolor=False, colors="w"
)
axx.set_title(f"latitude={lat}")
dcpy.plots.clean_axes(ax)
f.set_size_inches((8, 6))
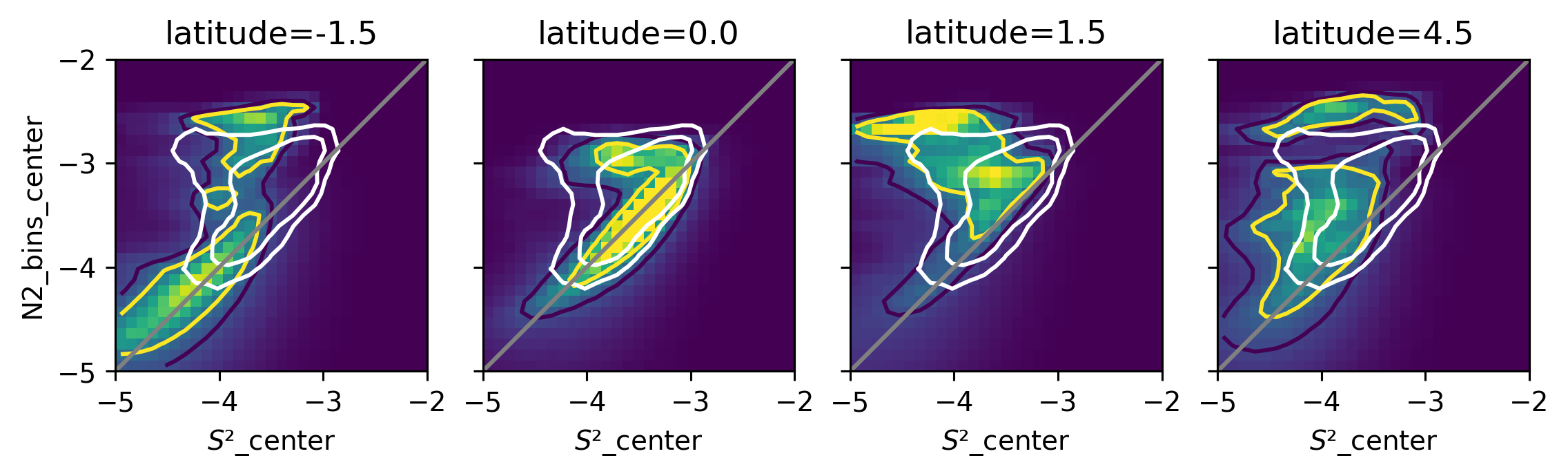
Here is TAO 0, 140 by itself
mixpods.plot_n2s2pdf(
tao_gridded.n2s2pdf.sel(enso_transition_phase="none"),
)
<matplotlib.contour.QuadContourSet at 0x2af9d92f38b0>
vs microstructure#
def integrate(da, dim):
import pandas as pd
bin_ = da[dim][0].data.item()
result = da.sum(dim) * bin_.length
for other_dim in result.dims:
maybe_bin = da[other_dim][0].data.item()
if isinstance(maybe_bin, pd.Interval):
result[other_dim] = pd.IntervalIndex(result[other_dim].data).mid.to_numpy()
return result
Show code cell source Hide code cell source
to_plot = {
"/les moor": moorings["0.0.-140.oct.mooring.month"].ds.load(),
"/les avg": avgs["0.0.-140.oct.average.month"].ds.load(),
"/tao oct": tao_gridded[["n2s2pdf_monthly"]]
.sel(month=10)
.rename({"n2s2pdf_monthly": "n2s2pdf"}),
}
to_plot.update(micro.to_dict())
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in true_divide
return func(*(_execute_task(a, cache) for a in args))
/glade/u/home/dcherian/miniconda3/envs/pump/lib/python3.10/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in log10
return func(*(_execute_task(a, cache) for a in args))
Show code cell source Hide code cell source
colors = {
"les avg": "magenta",
"les moor": "red",
"tao oct": "black",
"equix": "green",
"tropicheat": "darkgray",
"tiwe": "blue",
}
hdls = []
s2hdls = []
n2hdls = []
for name, node in to_plot.items():
if node.data_vars:
kwargs = dict(line_width=2, label=name[1:])
hdls.append(
mixpods.hvplot_n2s2pdf(
node["n2s2pdf"],
targets=(0.5,),
pcolor=False,
cmap=[colors[name[1:]]],
**kwargs
)
)
s2hdls.append(
integrate(node.n2s2pdf.squeeze(), "N2_bins").hvplot.line(
**kwargs, color=[colors[name[1:]]]
)
)
n2hdls.append(
integrate(node.n2s2pdf.squeeze(), "S2_bins").hvplot.line(
**kwargs, color=[colors[name[1:]]]
)
)
from functools import reduce
import operator
joint = (
reduce(operator.mul, hdls)
# .opts(frame_width=400, frame_height=400)
# .opts(xlabel="log S2", ylabel="log 4N2")
)
s2 = reduce(operator.mul, s2hdls)
n2 = reduce(operator.mul, n2hdls)
(
joint
<< n2.opts(show_legend=False) # frame_width=200, frame_height=400)
<< s2.opts(show_legend=False) ##.opts(frame_width=400, frame_height=200)
).opts(width=500)
(n2.opts(show_legend=True) + s2.opts(show_legend=True)).cols(1)
TODO#
Combined 5-day distributions: all longitudes
hour average for microstructure; maybe vertical scales too
LES Ri PDF with different subsampling z; match with Smyth & Moum (2013)
Add domain depth to catalog
TODO: Turbulence patterns#
moorings = moorings.assign(
{
"epsilon": lambda ds: (15 / 4 * ds.nu_sgs * ds.S2)
if "nu_sgs" in ds
else xr.DataArray([0])
}
)
del moorings.ds["epsilon"]
moorings
<xarray.Dataset>
Dimensions: (latitude: 4, enso_transition_phase: 1, N2_bins: 29,
S2_bins: 29)
Coordinates:
dz float64 0.5
longitude int64 140
* latitude (latitude) float64 -1.5 0.0 1.5 4.5
* enso_transition_phase (enso_transition_phase) object 'none'
* N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-...
* S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-...
bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107
variable <U7 'n2s2pdf'
Data variables:
n2s2pdf (latitude, enso_transition_phase, N2_bins, S2_bins) float64 ...datatree.DataTree
- z: 288
- time: 35659
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- z(z)float64-143.5 -143.0 -142.5 ... -0.5 0.0
- axis :
- Z
- long_name :
- depth
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- meters
- standard_name :
- depth
array([-143.5, -143. , -142.5, ..., -1. , -0.5, 0. ])
- time(time)datetime64[ns]1985-10-02T06:02:25.395800351 .....
- axis :
- T
- standard_name :
- time
- long_name :
- time since initialization
array(['1985-10-02T06:02:25.395800351', '1985-10-02T06:04:53.777924419', '1985-10-02T06:07:22.219675481', ..., '1985-11-05T20:47:51.086233894', '1985-11-05T20:49:09.047709473', '1985-11-05T20:50:26.981129554'], dtype='datetime64[ns]') - dz()float640.5
array(0.5)
- longitude()int64140
array(140)
- latitude()float644.5
array(4.5)
- enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
- units :
- s$^{-2}$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]])
- u(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_eastward_velocity
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - v(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_northward_velocity
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - w(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- inst. vert vel at one column
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - temp(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- inst. temp. at one column
- units :
- Celsius
- time :
- time
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - salt(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- inst. salinity at one column
- units :
- psu
- time :
- time
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - nu_sgs(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- subgrid turbulent viscosity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - kappa_sgs(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- subgrid turbulent diffusivity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - alpha()float640.0002976
- long_name :
- thermal expansion coeff., lin. eq. of state
array(0.00029755)
- beta()float64-0.0007386
- long_name :
- haline contraction coeff., lin. eq. of state
array(-0.00073863)
- T0()float6425.0
- long_name :
- reference temperature, lin eq. of state
array(25.)
- S0()float6435.25
- long_name :
- reference salinity, lin. eq. of state
array(35.25)
- rho0()float641.024e+03
- long_name :
- reference density, lin. eq. of state
array(1023.53)
- rho(time, z)float64dask.array<chunksize=(35659, 288), meta=np.ndarray>
- standard_name :
- sea_water_potential_density
Array Chunk Bytes 78.35 MiB 78.35 MiB Shape (35659, 288) (35659, 288) Count 20 Tasks 1 Chunks Type float64 numpy.ndarray - buoy(time, z)float64dask.array<chunksize=(35659, 288), meta=np.ndarray>
- standard_name :
- sea_water_buoyancy
Array Chunk Bytes 78.35 MiB 78.35 MiB Shape (35659, 288) (35659, 288) Count 22 Tasks 1 Chunks Type float64 numpy.ndarray - uz(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - vz(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - S2(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- $S^2$
- units :
- s$^{-2}$
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 31 Tasks 1 Chunks Type float32 numpy.ndarray - shear(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- |$u_z$|
- units :
- s$^{-1}$
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 30 Tasks 1 Chunks Type float32 numpy.ndarray - N2(time, z)float64dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- $N^2$
- units :
- s$^{-2}$
Array Chunk Bytes 78.35 MiB 78.35 MiB Shape (35659, 288) (35659, 288) Count 27 Tasks 1 Chunks Type float64 numpy.ndarray - shred2(time, z)float64dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- Reduced shear$^2$
- units :
- $s^{-2}$
Array Chunk Bytes 78.35 MiB 78.35 MiB Shape (35659, 288) (35659, 288) Count 58 Tasks 1 Chunks Type float64 numpy.ndarray - Ri(time, z)float64dask.array<chunksize=(35659, 288), meta=np.ndarray>
- long_name :
- Ri
- units :
Array Chunk Bytes 78.35 MiB 78.35 MiB Shape (35659, 288) (35659, 288) Count 57 Tasks 1 Chunks Type float64 numpy.ndarray - eucmax(time)float64-110.0 -110.0 ... -123.5 -123.5
- long_name :
- Depth of EUC max
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- m
- standard_name :
- depth
array([-110. , -110. , -110. , ..., -123.5, -123.5, -123.5])
- n2s2pdf(enso_transition_phase, N2_bins, S2_bins)float64dask.array<chunksize=(1, 29, 29), meta=np.ndarray>
- long_name :
- $P(S^2, 4N^2)$
Array Chunk Bytes 6.57 kiB 6.57 kiB Shape (1, 29, 29) (1, 29, 29) Count 79 Tasks 1 Chunks Type float64 numpy.ndarray - epsilon(time, z)float32dask.array<chunksize=(35659, 288), meta=np.ndarray>
Array Chunk Bytes 39.18 MiB 39.18 MiB Shape (35659, 288) (35659, 288) Count 40 Tasks 1 Chunks Type float32 numpy.ndarray
- type :
- DIABLO LES, processed virtual mooring
- title :
- ROMS_PSH_6HRLIN_4.5N140W_360x360x288_23NOV2021_mooring
- intake_esm_varname :
- ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa_sgs', 'alpha', 'beta', 'T0', 'S0', 'rho0')
- intake_esm_dataset_key :
- 4.5.140.oct.mooring.month
- z: 216
- time: 15342
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- z(z)float64-107.5 -107.0 -106.5 ... -0.5 0.0
- axis :
- Z
- long_name :
- depth
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- meters
- standard_name :
- depth
array([-107.5, -107. , -106.5, ..., -1. , -0.5, 0. ])
- time(time)datetime64[ns]1985-10-02T06:01:13.845228446 .....
- axis :
- T
- standard_name :
- time
- long_name :
- time since initialization
array(['1985-10-02T06:01:13.845228446', '1985-10-02T06:04:57.167508625', '1985-10-02T06:08:36.606096207', ..., '1985-11-05T20:50:22.724547005', '1985-11-05T20:53:32.482677670', '1985-11-05T20:56:42.407805497'], dtype='datetime64[ns]') - dz()float640.5
array(0.5)
- longitude()int64140
array(140)
- latitude()float640.0
array(0.)
- enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
- units :
- s$^{-2}$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]])
- u(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_eastward_velocity
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - v(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_northward_velocity
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - w(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- inst. vert vel at one column
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - temp(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- inst. temp. at one column
- units :
- Celsius
- time :
- time
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - salt(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- inst. salinity at one column
- units :
- psu
- time :
- time
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - nu_sgs(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- subgrid turbulent viscosity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - kappa_sgs(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- subgrid turbulent diffusivity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - alpha()float640.0002976
- long_name :
- thermal expansion coeff., lin. eq. of state
array(0.00029755)
- beta()float64-0.0007386
- long_name :
- haline contraction coeff., lin. eq. of state
array(-0.00073863)
- T0()float6425.0
- long_name :
- reference temperature, lin eq. of state
array(25.)
- S0()float6435.25
- long_name :
- reference salinity, lin. eq. of state
array(35.25)
- rho0()float641.024e+03
- long_name :
- reference density, lin. eq. of state
array(1023.53)
- rho(time, z)float64dask.array<chunksize=(15342, 216), meta=np.ndarray>
- standard_name :
- sea_water_potential_density
Array Chunk Bytes 25.28 MiB 25.28 MiB Shape (15342, 216) (15342, 216) Count 20 Tasks 1 Chunks Type float64 numpy.ndarray - buoy(time, z)float64dask.array<chunksize=(15342, 216), meta=np.ndarray>
- standard_name :
- sea_water_buoyancy
Array Chunk Bytes 25.28 MiB 25.28 MiB Shape (15342, 216) (15342, 216) Count 22 Tasks 1 Chunks Type float64 numpy.ndarray - uz(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - vz(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - S2(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- $S^2$
- units :
- s$^{-2}$
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 31 Tasks 1 Chunks Type float32 numpy.ndarray - shear(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- |$u_z$|
- units :
- s$^{-1}$
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 30 Tasks 1 Chunks Type float32 numpy.ndarray - N2(time, z)float64dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- $N^2$
- units :
- s$^{-2}$
Array Chunk Bytes 25.28 MiB 25.28 MiB Shape (15342, 216) (15342, 216) Count 27 Tasks 1 Chunks Type float64 numpy.ndarray - shred2(time, z)float64dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- Reduced shear$^2$
- units :
- $s^{-2}$
Array Chunk Bytes 25.28 MiB 25.28 MiB Shape (15342, 216) (15342, 216) Count 58 Tasks 1 Chunks Type float64 numpy.ndarray - Ri(time, z)float64dask.array<chunksize=(15342, 216), meta=np.ndarray>
- long_name :
- Ri
- units :
Array Chunk Bytes 25.28 MiB 25.28 MiB Shape (15342, 216) (15342, 216) Count 57 Tasks 1 Chunks Type float64 numpy.ndarray - eucmax(time)float64nan nan nan ... -104.0 -104.0
- long_name :
- Depth of EUC max
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- m
- standard_name :
- depth
array([ nan, nan, nan, ..., -104., -104., -104.])
- n2s2pdf(enso_transition_phase, N2_bins, S2_bins)float64dask.array<chunksize=(1, 29, 29), meta=np.ndarray>
- long_name :
- $P(S^2, 4N^2)$
Array Chunk Bytes 6.57 kiB 6.57 kiB Shape (1, 29, 29) (1, 29, 29) Count 79 Tasks 1 Chunks Type float64 numpy.ndarray - epsilon(time, z)float32dask.array<chunksize=(15342, 216), meta=np.ndarray>
Array Chunk Bytes 12.64 MiB 12.64 MiB Shape (15342, 216) (15342, 216) Count 40 Tasks 1 Chunks Type float32 numpy.ndarray
- type :
- DIABLO LES, processed virtual mooring
- title :
- ROMS_PSH_6HRLIN_0N140W_360x360x21_22OCT2020_mooring
- intake_esm_varname :
- ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa_sgs', 'alpha', 'beta', 'T0', 'S0', 'rho0')
- intake_esm_dataset_key :
- 0.0.140.oct.mooring.month
- z: 288
- time: 32515
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- z(z)float64-143.5 -143.0 -142.5 ... -0.5 0.0
- axis :
- Z
- long_name :
- depth
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- meters
- standard_name :
- depth
array([-143.5, -143. , -142.5, ..., -1. , -0.5, 0. ])
- time(time)datetime64[ns]1985-10-02T06:02:29.072796441 .....
- axis :
- T
- standard_name :
- time
- long_name :
- time since initialization
array(['1985-10-02T06:02:29.072796441', '1985-10-02T06:05:01.175053110', '1985-10-02T06:07:33.294448443', ..., '1985-11-05T20:36:30.017604646', '1985-11-05T20:38:26.898830198', '1985-11-05T20:40:23.801460663'], dtype='datetime64[ns]') - dz()float640.5
array(0.5)
- longitude()int64140
array(140)
- latitude()float64-1.5
array(-1.5)
- enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
- units :
- s$^{-2}$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]])
- u(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_eastward_velocity
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - v(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_northward_velocity
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - w(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- inst. vert vel at one column
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - temp(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- inst. temp. at one column
- units :
- Celsius
- time :
- time
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - salt(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- inst. salinity at one column
- units :
- psu
- time :
- time
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - nu_sgs(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- subgrid turbulent viscosity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - kappa_sgs(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- subgrid turbulent diffusivity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - alpha()float640.0002976
- long_name :
- thermal expansion coeff., lin. eq. of state
array(0.00029755)
- beta()float64-0.0007386
- long_name :
- haline contraction coeff., lin. eq. of state
array(-0.00073863)
- T0()float6425.0
- long_name :
- reference temperature, lin eq. of state
array(25.)
- S0()float6435.25
- long_name :
- reference salinity, lin. eq. of state
array(35.25)
- rho0()float641.024e+03
- long_name :
- reference density, lin. eq. of state
array(1023.53)
- rho(time, z)float64dask.array<chunksize=(32515, 288), meta=np.ndarray>
- standard_name :
- sea_water_potential_density
Array Chunk Bytes 71.44 MiB 71.44 MiB Shape (32515, 288) (32515, 288) Count 20 Tasks 1 Chunks Type float64 numpy.ndarray - buoy(time, z)float64dask.array<chunksize=(32515, 288), meta=np.ndarray>
- standard_name :
- sea_water_buoyancy
Array Chunk Bytes 71.44 MiB 71.44 MiB Shape (32515, 288) (32515, 288) Count 22 Tasks 1 Chunks Type float64 numpy.ndarray - uz(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - vz(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - S2(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- $S^2$
- units :
- s$^{-2}$
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 31 Tasks 1 Chunks Type float32 numpy.ndarray - shear(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- |$u_z$|
- units :
- s$^{-1}$
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 30 Tasks 1 Chunks Type float32 numpy.ndarray - N2(time, z)float64dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- $N^2$
- units :
- s$^{-2}$
Array Chunk Bytes 71.44 MiB 71.44 MiB Shape (32515, 288) (32515, 288) Count 27 Tasks 1 Chunks Type float64 numpy.ndarray - shred2(time, z)float64dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- Reduced shear$^2$
- units :
- $s^{-2}$
Array Chunk Bytes 71.44 MiB 71.44 MiB Shape (32515, 288) (32515, 288) Count 58 Tasks 1 Chunks Type float64 numpy.ndarray - Ri(time, z)float64dask.array<chunksize=(32515, 288), meta=np.ndarray>
- long_name :
- Ri
- units :
Array Chunk Bytes 71.44 MiB 71.44 MiB Shape (32515, 288) (32515, 288) Count 57 Tasks 1 Chunks Type float64 numpy.ndarray - eucmax(time)float64-128.5 -128.0 ... -130.5 -131.0
- long_name :
- Depth of EUC max
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- m
- standard_name :
- depth
array([-128.5, -128. , -128.5, ..., -130. , -130.5, -131. ])
- n2s2pdf(enso_transition_phase, N2_bins, S2_bins)float64dask.array<chunksize=(1, 29, 29), meta=np.ndarray>
- long_name :
- $P(S^2, 4N^2)$
Array Chunk Bytes 6.57 kiB 6.57 kiB Shape (1, 29, 29) (1, 29, 29) Count 79 Tasks 1 Chunks Type float64 numpy.ndarray - epsilon(time, z)float32dask.array<chunksize=(32515, 288), meta=np.ndarray>
Array Chunk Bytes 35.72 MiB 35.72 MiB Shape (32515, 288) (32515, 288) Count 40 Tasks 1 Chunks Type float32 numpy.ndarray
- type :
- DIABLO LES, processed virtual mooring
- title :
- ROMS_PSH_6HRLIN_1.5S140W_360x360x288_23NOV2021_mooring
- intake_esm_varname :
- ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa_sgs', 'alpha', 'beta', 'T0', 'S0', 'rho0')
- intake_esm_dataset_key :
- -1.5.140.oct.mooring.month
- z: 288
- time: 38132
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- z(z)float64-143.5 -143.0 -142.5 ... -0.5 0.0
- axis :
- Z
- long_name :
- depth
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- meters
- standard_name :
- depth
array([-143.5, -143. , -142.5, ..., -1. , -0.5, 0. ])
- time(time)datetime64[ns]1985-10-02T06:01:42.416460963 .....
- axis :
- T
- standard_name :
- time
- long_name :
- time since initialization
array(['1985-10-02T06:01:42.416460963', '1985-10-02T06:03:25.828030185', '1985-10-02T06:05:08.177447770', ..., '1985-11-05T17:09:43.639123264', '1985-11-05T17:10:49.296434130', '1985-11-05T17:11:54.853028559'], dtype='datetime64[ns]') - dz()float640.5
array(0.5)
- longitude()int64140
array(140)
- latitude()float641.5
array(1.5)
- enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
- units :
- s$^{-2}$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]])
- u(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_eastward_velocity
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - v(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- inst. horiz. vel. at one column
- units :
- meter second-1
- time :
- time
- standard_name :
- sea_water_northward_velocity
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - w(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- inst. vert vel at one column
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - temp(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- inst. temp. at one column
- units :
- Celsius
- time :
- time
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - salt(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- inst. salinity at one column
- units :
- psu
- time :
- time
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - nu_sgs(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- subgrid turbulent viscosity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - kappa_sgs(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- subgrid turbulent diffusivity
- units :
- meter2 second-1
- time :
- time
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 7 Tasks 1 Chunks Type float32 numpy.ndarray - alpha()float640.0002976
- long_name :
- thermal expansion coeff., lin. eq. of state
array(0.00029755)
- beta()float64-0.0007386
- long_name :
- haline contraction coeff., lin. eq. of state
array(-0.00073863)
- T0()float6425.0
- long_name :
- reference temperature, lin eq. of state
array(25.)
- S0()float6435.25
- long_name :
- reference salinity, lin. eq. of state
array(35.25)
- rho0()float641.024e+03
- long_name :
- reference density, lin. eq. of state
array(1023.53)
- rho(time, z)float64dask.array<chunksize=(38132, 288), meta=np.ndarray>
- standard_name :
- sea_water_potential_density
Array Chunk Bytes 83.79 MiB 83.79 MiB Shape (38132, 288) (38132, 288) Count 20 Tasks 1 Chunks Type float64 numpy.ndarray - buoy(time, z)float64dask.array<chunksize=(38132, 288), meta=np.ndarray>
- standard_name :
- sea_water_buoyancy
Array Chunk Bytes 83.79 MiB 83.79 MiB Shape (38132, 288) (38132, 288) Count 22 Tasks 1 Chunks Type float64 numpy.ndarray - uz(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - vz(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 13 Tasks 1 Chunks Type float32 numpy.ndarray - S2(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- $S^2$
- units :
- s$^{-2}$
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 31 Tasks 1 Chunks Type float32 numpy.ndarray - shear(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- |$u_z$|
- units :
- s$^{-1}$
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 30 Tasks 1 Chunks Type float32 numpy.ndarray - N2(time, z)float64dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- $N^2$
- units :
- s$^{-2}$
Array Chunk Bytes 83.79 MiB 83.79 MiB Shape (38132, 288) (38132, 288) Count 27 Tasks 1 Chunks Type float64 numpy.ndarray - shred2(time, z)float64dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- Reduced shear$^2$
- units :
- $s^{-2}$
Array Chunk Bytes 83.79 MiB 83.79 MiB Shape (38132, 288) (38132, 288) Count 58 Tasks 1 Chunks Type float64 numpy.ndarray - Ri(time, z)float64dask.array<chunksize=(38132, 288), meta=np.ndarray>
- long_name :
- Ri
- units :
Array Chunk Bytes 83.79 MiB 83.79 MiB Shape (38132, 288) (38132, 288) Count 57 Tasks 1 Chunks Type float64 numpy.ndarray - eucmax(time)float64-125.0 -125.0 ... -111.5 -111.5
- long_name :
- Depth of EUC max
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- m
- standard_name :
- depth
array([-125. , -125. , -124.5, ..., -110.5, -111.5, -111.5])
- n2s2pdf(enso_transition_phase, N2_bins, S2_bins)float64dask.array<chunksize=(1, 29, 29), meta=np.ndarray>
- long_name :
- $P(S^2, 4N^2)$
Array Chunk Bytes 6.57 kiB 6.57 kiB Shape (1, 29, 29) (1, 29, 29) Count 79 Tasks 1 Chunks Type float64 numpy.ndarray - epsilon(time, z)float32dask.array<chunksize=(38132, 288), meta=np.ndarray>
Array Chunk Bytes 41.89 MiB 41.89 MiB Shape (38132, 288) (38132, 288) Count 40 Tasks 1 Chunks Type float32 numpy.ndarray
- type :
- DIABLO LES, processed virtual mooring
- title :
- ROMS_PSH_6HRLIN_1.5N140W_360x360x288_5OCT2021_mooring
- intake_esm_varname :
- ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa_sgs', 'alpha', 'beta', 'T0', 'S0', 'rho0')
- intake_esm_dataset_key :
- 1.5.140.oct.mooring.month
<xarray.Dataset> Dimensions: (z: 288, time: 35659, enso_transition_phase: 1, N2_bins: 29, S2_bins: 29) Coordinates: * z (z) float64 -143.5 -143.0 -142.5 ... -1.0 -0.5 0.0 * time (time) datetime64[ns] 1985-10-02T06:02:25.39580035... dz float64 0.5 longitude int64 140 latitude float64 4.5 * enso_transition_phase (enso_transition_phase) object 'none' * N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-... * S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-... bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107 Data variables: (12/24) u (time, z) float32 dask.array<chunksize=(35659, 288), meta=np.ndarray> v (time, z) float32 dask.array<chunksize=(35659, 288), meta=np.ndarray> w (time, z) float32 dask.array<chunksize=(35659, 288), meta=np.ndarray> temp (time, z) float32 dask.array<chunksize=(35659, 288), meta=np.ndarray> salt (time, z) float32 dask.array<chunksize=(35659, 288), meta=np.ndarray> nu_sgs (time, z) float32 dask.array<chunksize=(35659, 288), meta=np.ndarray> ... ... N2 (time, z) float64 dask.array<chunksize=(35659, 288), meta=np.ndarray> shred2 (time, z) float64 dask.array<chunksize=(35659, 288), meta=np.ndarray> Ri (time, z) float64 dask.array<chunksize=(35659, 288), meta=np.ndarray> eucmax (time) float64 -110.0 -110.0 -110.0 ... -123.5 -123.5 n2s2pdf (enso_transition_phase, N2_bins, S2_bins) float64 dask.array<chunksize=(1, 29, 29), meta=np.ndarray> epsilon (time, z) float32 dask.array<chunksize=(35659, 288), meta=np.ndarray> Attributes: type: DIABLO LES, processed virtual mooring title: ROMS_PSH_6HRLIN_4.5N140W_360x360x288_23NOV2021_m... intake_esm_varname: ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa... intake_esm_dataset_key: 4.5.140.oct.mooring.month4.5.140.oct.mooring.month<xarray.Dataset> Dimensions: (z: 216, time: 15342, enso_transition_phase: 1, N2_bins: 29, S2_bins: 29) Coordinates: * z (z) float64 -107.5 -107.0 -106.5 ... -1.0 -0.5 0.0 * time (time) datetime64[ns] 1985-10-02T06:01:13.84522844... dz float64 0.5 longitude int64 140 latitude float64 0.0 * enso_transition_phase (enso_transition_phase) object 'none' * N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-... * S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-... bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107 Data variables: (12/24) u (time, z) float32 dask.array<chunksize=(15342, 216), meta=np.ndarray> v (time, z) float32 dask.array<chunksize=(15342, 216), meta=np.ndarray> w (time, z) float32 dask.array<chunksize=(15342, 216), meta=np.ndarray> temp (time, z) float32 dask.array<chunksize=(15342, 216), meta=np.ndarray> salt (time, z) float32 dask.array<chunksize=(15342, 216), meta=np.ndarray> nu_sgs (time, z) float32 dask.array<chunksize=(15342, 216), meta=np.ndarray> ... ... N2 (time, z) float64 dask.array<chunksize=(15342, 216), meta=np.ndarray> shred2 (time, z) float64 dask.array<chunksize=(15342, 216), meta=np.ndarray> Ri (time, z) float64 dask.array<chunksize=(15342, 216), meta=np.ndarray> eucmax (time) float64 nan nan nan ... -104.0 -104.0 -104.0 n2s2pdf (enso_transition_phase, N2_bins, S2_bins) float64 dask.array<chunksize=(1, 29, 29), meta=np.ndarray> epsilon (time, z) float32 dask.array<chunksize=(15342, 216), meta=np.ndarray> Attributes: type: DIABLO LES, processed virtual mooring title: ROMS_PSH_6HRLIN_0N140W_360x360x21_22OCT2020_mooring intake_esm_varname: ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa... intake_esm_dataset_key: 0.0.140.oct.mooring.month0.0.140.oct.mooring.month<xarray.Dataset> Dimensions: (z: 288, time: 32515, enso_transition_phase: 1, N2_bins: 29, S2_bins: 29) Coordinates: * z (z) float64 -143.5 -143.0 -142.5 ... -1.0 -0.5 0.0 * time (time) datetime64[ns] 1985-10-02T06:02:29.07279644... dz float64 0.5 longitude int64 140 latitude float64 -1.5 * enso_transition_phase (enso_transition_phase) object 'none' * N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-... * S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-... bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107 Data variables: (12/24) u (time, z) float32 dask.array<chunksize=(32515, 288), meta=np.ndarray> v (time, z) float32 dask.array<chunksize=(32515, 288), meta=np.ndarray> w (time, z) float32 dask.array<chunksize=(32515, 288), meta=np.ndarray> temp (time, z) float32 dask.array<chunksize=(32515, 288), meta=np.ndarray> salt (time, z) float32 dask.array<chunksize=(32515, 288), meta=np.ndarray> nu_sgs (time, z) float32 dask.array<chunksize=(32515, 288), meta=np.ndarray> ... ... N2 (time, z) float64 dask.array<chunksize=(32515, 288), meta=np.ndarray> shred2 (time, z) float64 dask.array<chunksize=(32515, 288), meta=np.ndarray> Ri (time, z) float64 dask.array<chunksize=(32515, 288), meta=np.ndarray> eucmax (time) float64 -128.5 -128.0 -128.5 ... -130.5 -131.0 n2s2pdf (enso_transition_phase, N2_bins, S2_bins) float64 dask.array<chunksize=(1, 29, 29), meta=np.ndarray> epsilon (time, z) float32 dask.array<chunksize=(32515, 288), meta=np.ndarray> Attributes: type: DIABLO LES, processed virtual mooring title: ROMS_PSH_6HRLIN_1.5S140W_360x360x288_23NOV2021_m... intake_esm_varname: ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa... intake_esm_dataset_key: -1.5.140.oct.mooring.month-1.5.140.oct.mooring.month<xarray.Dataset> Dimensions: (z: 288, time: 38132, enso_transition_phase: 1, N2_bins: 29, S2_bins: 29) Coordinates: * z (z) float64 -143.5 -143.0 -142.5 ... -1.0 -0.5 0.0 * time (time) datetime64[ns] 1985-10-02T06:01:42.41646096... dz float64 0.5 longitude int64 140 latitude float64 1.5 * enso_transition_phase (enso_transition_phase) object 'none' * N2_bins (N2_bins) object (-5.0, -4.896551724137931] ... (-... * S2_bins (S2_bins) object (-5.0, -4.896551724137931] ... (-... bin_areas (N2_bins, S2_bins) float64 0.0107 0.0107 ... 0.0107 Data variables: (12/24) u (time, z) float32 dask.array<chunksize=(38132, 288), meta=np.ndarray> v (time, z) float32 dask.array<chunksize=(38132, 288), meta=np.ndarray> w (time, z) float32 dask.array<chunksize=(38132, 288), meta=np.ndarray> temp (time, z) float32 dask.array<chunksize=(38132, 288), meta=np.ndarray> salt (time, z) float32 dask.array<chunksize=(38132, 288), meta=np.ndarray> nu_sgs (time, z) float32 dask.array<chunksize=(38132, 288), meta=np.ndarray> ... ... N2 (time, z) float64 dask.array<chunksize=(38132, 288), meta=np.ndarray> shred2 (time, z) float64 dask.array<chunksize=(38132, 288), meta=np.ndarray> Ri (time, z) float64 dask.array<chunksize=(38132, 288), meta=np.ndarray> eucmax (time) float64 -125.0 -125.0 -124.5 ... -111.5 -111.5 n2s2pdf (enso_transition_phase, N2_bins, S2_bins) float64 dask.array<chunksize=(1, 29, 29), meta=np.ndarray> epsilon (time, z) float32 dask.array<chunksize=(38132, 288), meta=np.ndarray> Attributes: type: DIABLO LES, processed virtual mooring title: ROMS_PSH_6HRLIN_1.5N140W_360x360x288_5OCT2021_mo... intake_esm_varname: ('u', 'v', 'w', 'temp', 'salt', 'nu_sgs', 'kappa... intake_esm_dataset_key: 1.5.140.oct.mooring.month1.5.140.oct.mooring.month- latitude: 4
- enso_transition_phase: 1
- N2_bins: 29
- S2_bins: 29
- dz()float640.5
array(0.5)
- longitude()int64140
array(140)
- latitude(latitude)float64-1.5 0.0 1.5 4.5
array([-1.5, 0. , 1.5, 4.5])
- enso_transition_phase(enso_transition_phase)object'none'
array(['none'], dtype=object)
- N2_bins(N2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} 4N^2$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - S2_bins(S2_bins)object(-5.0, -4.896551724137931] ... (...
- long_name :
- log$_{10} S^2$
- units :
- s$^{-2}$
array([Interval(-5.0, -4.896551724137931, closed='right'), Interval(-4.896551724137931, -4.793103448275862, closed='right'), Interval(-4.793103448275862, -4.689655172413793, closed='right'), Interval(-4.689655172413793, -4.586206896551724, closed='right'), Interval(-4.586206896551724, -4.482758620689655, closed='right'), Interval(-4.482758620689655, -4.379310344827586, closed='right'), Interval(-4.379310344827586, -4.275862068965517, closed='right'), Interval(-4.275862068965517, -4.172413793103448, closed='right'), Interval(-4.172413793103448, -4.068965517241379, closed='right'), Interval(-4.068965517241379, -3.9655172413793105, closed='right'), Interval(-3.9655172413793105, -3.862068965517241, closed='right'), Interval(-3.862068965517241, -3.7586206896551726, closed='right'), Interval(-3.7586206896551726, -3.655172413793103, closed='right'), Interval(-3.655172413793103, -3.5517241379310347, closed='right'), Interval(-3.5517241379310347, -3.4482758620689653, closed='right'), Interval(-3.4482758620689653, -3.344827586206897, closed='right'), Interval(-3.344827586206897, -3.2413793103448274, closed='right'), Interval(-3.2413793103448274, -3.137931034482759, closed='right'), Interval(-3.137931034482759, -3.0344827586206895, closed='right'), Interval(-3.0344827586206895, -2.9310344827586206, closed='right'), Interval(-2.9310344827586206, -2.8275862068965516, closed='right'), Interval(-2.8275862068965516, -2.7241379310344827, closed='right'), Interval(-2.7241379310344827, -2.6206896551724137, closed='right'), Interval(-2.6206896551724137, -2.5172413793103448, closed='right'), Interval(-2.5172413793103448, -2.413793103448276, closed='right'), Interval(-2.413793103448276, -2.310344827586207, closed='right'), Interval(-2.310344827586207, -2.206896551724138, closed='right'), Interval(-2.206896551724138, -2.103448275862069, closed='right'), Interval(-2.103448275862069, -2.0, closed='right')], dtype=object) - bin_areas(N2_bins, S2_bins)float640.0107 0.0107 ... 0.0107 0.0107
- long_name :
- log$_{10} 4N^2$
array([[0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, ... 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155], [0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155, 0.01070155]]) - variable()<U7'n2s2pdf'
array('n2s2pdf', dtype='<U7')
- n2s2pdf(latitude, enso_transition_phase, N2_bins, S2_bins)float640.222 0.214 0.2125 ... 0.0 0.0 0.0
array([[[[2.22043869e-01, 2.13992239e-01, 2.12499384e-01, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [2.87876340e-01, 2.77915813e-01, 2.62032812e-01, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [4.22673830e-01, 4.14475362e-01, 3.71574128e-01, ..., 2.44730374e-05, 0.00000000e+00, 0.00000000e+00], ..., [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], [[[4.09186509e-02, 4.82869554e-02, 4.98588603e-02, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [5.23149618e-02, 6.11569272e-02, 6.40551269e-02, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [6.68059606e-02, 7.81040275e-02, 8.29179864e-02, ..., ... 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]], [[[1.21128343e-01, 1.23521113e-01, 1.30699423e-01, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.37652001e-01, 1.46455589e-01, 1.48126013e-01, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [1.51421715e-01, 1.65326870e-01, 1.73633845e-01, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], ..., [1.35439817e-04, 9.02932114e-05, 2.25733028e-04, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 4.51466057e-05, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00], [0.00000000e+00, 0.00000000e+00, 0.00000000e+00, ..., 0.00000000e+00, 0.00000000e+00, 0.00000000e+00]]]])
moorings.ds["epsilon"] = (
to_dataset(extract(clear_root(moorings), "epsilon"), dim="latitude")
.to_array()
.squeeze()
)
dz
longitude
latitude
enso_transition_phase
N2_bins
S2_bins
bin_areas
variable
n2s2pdf
avgs.ds["epsilon"] = (
to_dataset(extract(clear_root(avgs), "epsilon"), dim="latitude")
.to_array()
.squeeze()
)
dz
longitude
latitude
enso_transition_phase
N2_bins
S2_bins
bin_areas
variable
n2s2pdf
Build Catalog#
Using ecgtools
Dan:
(100 W is grid point “1402” and 165 W is grid point “102”; 140W is “602” first date “25MAY1985” indicates first simulated date; second date 24NOV2021 is the nominal date I started the runs):
import glob
import pathlib
import ecgtools
import intake
import intake_esm
import xarray as xr
root = (
"/glade/p/cgd/oce/people/dwhitt/TPOS/"
"tpos_LES_runs_setup_scripts/tpos-DIABLO/diablo_2.0/post_process/diablo_analysis"
)
def parse_les_file(path):
path = pathlib.Path(path)
split = path.stem.split("_")
# print(split)
# print(path)
if split[3] == "242":
length = "5-day"
kind = "average"
longitude = int(-165 + (int(split[4]) - 102) * 0.05)
latitude = 0
else:
length = "month"
kind = "mooring" if "mooring" in split[-1] else "average"
longitude = -1 * int(split[3][-4:-1])
latitude = float(split[3][:-5]) * (-1 if "S" in split[3] else 1)
with xr.open_dataset(path) as ds:
info = {
"length": length,
"kind": kind,
"longitude": longitude,
"latitude": latitude,
"month": "may" if "MAY" in split[-2] else "oct",
"path": path,
"variables": [k for k in ds],
}
return info
parse_les_file(f"{root}/ROMS_PSH_6HRLIN_0N140W_360x360x216_22OCT2020.nc")
{'length': 'month',
'kind': 'average',
'longitude': -140,
'latitude': 0.0,
'month': 'oct',
'path': PosixPath('/glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_runs_setup_scripts/tpos-DIABLO/diablo_2.0/post_process/diablo_analysis/ROMS_PSH_6HRLIN_0N140W_360x360x216_22OCT2020.nc'),
'variables': ['ume',
'vme',
'tempme',
'saltme',
'urms',
'vrms',
'wrms',
'temprms',
'saltrms',
'uw',
'vw',
'saltw',
'tempw',
'nududz',
'nudvdz',
'kappadsdz',
'kappadtdz',
'dTdtSOLAR',
'dTdtRESTORE',
'dTdtFORCE',
'dUdtRESTORE',
'dUdtFORCE',
'dVdtRESTORE',
'dVdtFORCE',
'epsilon',
'N2',
'S2',
'RIG',
'nududztop',
'nudvdztop',
'kappadsdztop',
'kappadtdztop',
'nududzbot',
'nudvdzbot',
'kappadsdzbot',
'kappadtdzbot',
'alpha',
'beta',
'T0',
'S0',
'rho0']}
builder = ecgtools.Builder(
root,
exclude_patterns=[
"*test*",
"*irene_*",
"*spectra*",
"*fixedeps*",
"*timeavg*",
"*0N140W*5OCT202*",
"*0N140W*20OCT202*",
"*0N140W*29OCT202*",
"*6mavg*",
"*54x54*",
],
njobs=-1,
)
builder.build(parse_les_file)
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=-1)]: Done 1 out of 1 | elapsed: 0.9s finished
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=-1)]: Done 14 tasks | elapsed: 3.2s
[Parallel(n_jobs=-1)]: Done 36 out of 36 | elapsed: 15.7s finished
Builder(root_path=PosixPath('/glade/p/cgd/oce/people/dwhitt/TPOS/tpos_LES_runs_setup_scripts/tpos-DIABLO/diablo_2.0/post_process/diablo_analysis'), extension='.nc', depth=0, exclude_patterns=['*test*', '*irene_*', '*spectra*', '*fixedeps*', '*timeavg*', '*0N140W*5OCT202*', '*0N140W*20OCT202*', '*0N140W*29OCT202*', '*6mavg*', '*54x54*'], njobs=-1)
Summarize catalog#
builder.df.drop("variables", axis=1).groupby(
["length", "kind", "latitude", "longitude", "month"]
).count()
| path | |||||
|---|---|---|---|---|---|
| length | kind | latitude | longitude | month | |
| 5-day | average | 0.0 | -165 | may | 1 |
| oct | 1 | ||||
| -160 | may | 1 | |||
| oct | 1 | ||||
| -155 | may | 1 | |||
| oct | 1 | ||||
| -150 | may | 1 | |||
| oct | 1 | ||||
| -145 | may | 1 | |||
| oct | 1 | ||||
| -140 | may | 1 | |||
| -135 | may | 1 | |||
| oct | 1 | ||||
| -130 | may | 1 | |||
| oct | 1 | ||||
| -125 | may | 1 | |||
| oct | 1 | ||||
| -120 | may | 1 | |||
| oct | 1 | ||||
| -115 | may | 1 | |||
| oct | 1 | ||||
| -110 | may | 1 | |||
| oct | 1 | ||||
| -105 | may | 1 | |||
| oct | 1 | ||||
| -100 | may | 1 | |||
| oct | 1 | ||||
| month | average | -1.5 | -140 | oct | 1 |
| 0.0 | -140 | oct | 1 | ||
| 1.5 | -140 | oct | 1 | ||
| 3.0 | -140 | oct | 1 | ||
| 4.5 | -140 | oct | 1 | ||
| mooring | -1.5 | -140 | oct | 1 | |
| 0.0 | -140 | oct | 1 | ||
| 1.5 | -140 | oct | 1 | ||
| 4.5 | -140 | oct | 1 |
builder.save(
"../catalogs/pump-les-catalog.csv",
path_column_name="path",
variable_column_name="variables",
data_format="netcdf",
groupby_attrs=["latitude", "longitude", "month", "kind", "length"],
aggregations=[
{"type": "union", "attribute_name": "variables"},
{"type": "join_new", "attribute_name": "latitude"},
{"type": "join_new", "attribute_name": "longitude"},
],
)
Saved catalog location: ../catalogs/pump-les-catalog.json and ../catalogs/pump-les-catalog.csv
import ast
from datatree import DataTree
catalog = intake.open_esm_datastore(
"../catalogs/pump-les-catalog.json",
csv_kwargs={"converters": {"variables": ast.literal_eval}},
)
catalog.df
dataset_dict = catalog.to_dataset_dict(cdf_kwargs={})
--> The keys in the returned dictionary of datasets are constructed as follows:
'latitude.longitude.month.kind.length'
100.00% [36/36 00:28<00:00]
tree = DataTree.from_dict(dataset_dict)
tree["-140.may"].ds
<xarray.Dataset>
Dimensions: (z: 288, time: 1313, longitude: 1, month: 1)
Coordinates:
* z (z) float64 -143.5 -143.0 -142.5 ... -1.0 -0.5 9.969e+36
* time (time) datetime64[ns] 1985-05-25T03:00:00 ... 1985-05-30T00...
* month (month) <U3 'may'
* longitude (longitude) int64 -140
Data variables: (12/41)
ume (longitude, month, time, z) float32 dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
vme (longitude, month, time, z) float32 dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
tempme (longitude, month, time, z) float32 dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
saltme (longitude, month, time, z) float32 dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
urms (longitude, month, time, z) float32 dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
vrms (longitude, month, time, z) float32 dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
... ...
kappadtdzbot (longitude, month, time) float32 dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
alpha (longitude, month) float64 0.0002976
beta (longitude, month) float64 -0.0007386
T0 (longitude, month) float64 25.0
S0 (longitude, month) float64 35.25
rho0 (longitude, month) float64 1.024e+03
Attributes:
type: DIABLO LES, processed means
title: ROMS_PSH_6HRLIN_0N140W_360x360x288_5OCT2021
history: Tue Dec 28 09:09:16 2021: ncatted -O -a units,ti...
NCO: netCDF Operators version 4.9.5 (Homepage = http:...
intake_esm_varname: ('ume', 'vme', 'tempme', 'saltme', 'urms', 'vrms...
intake_esm_dataset_key: -140.mayxarray.Dataset
- z: 288
- time: 1313
- longitude: 1
- month: 1
- z(z)float64-143.5 -143.0 ... -0.5 9.969e+36
- long_name :
- depth
- valid_min :
- -100000.0
- valid_max :
- 100000.0
- positive :
- up
- units :
- meters
- standard_name :
- depth
array([-1.43500e+02, -1.43000e+02, -1.42500e+02, ..., -1.00000e+00, -5.00000e-01, 9.96921e+36]) - time(time)datetime64[ns]1985-05-25T03:00:00 ... 1985-05-...
- long_name :
- time since initialization
array(['1985-05-25T03:00:00.000000000', '1985-05-25T03:05:05.541287497', '1985-05-25T03:10:12.823093595', ..., '1985-05-30T00:02:11.062769461', '1985-05-30T00:07:41.818673684', '1985-05-30T00:13:12.612216141'], dtype='datetime64[ns]') - month(month)<U3'may'
array(['may'], dtype='<U3')
- longitude(longitude)int64-140
array([-140])
- ume(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- horiz. avg. u-momentum
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - vme(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- horiz. avg. v-momentum
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - tempme(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- horiz. avg. temperature
- units :
- Celsius
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - saltme(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- horiz. avg. salinity
- units :
- psu
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - urms(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- root-mean-square zonal momentum
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - vrms(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- root-mean-square merid momentum
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - wrms(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- root-mean-square vert momentum
- units :
- meter second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - temprms(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- root-mean-square temperature
- units :
- Celsius
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - saltrms(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- salinity
- units :
- psu
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - uw(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. adv. flux zonal momentum
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - vw(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. adv. flux merid momentum
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - saltw(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. adv. flux salt
- units :
- meter second-1 psu
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - tempw(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. adv. flux temp
- units :
- meter second-1 Celcius
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - nududz(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. visc. flux u-momentum
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - nudvdz(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. visc. flux v-momentum
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - kappadsdz(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. diff. flux salt
- units :
- meter second-1 psu
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - kappadtdz(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- vert. diff. flux temp
- units :
- meter second-1 Celcius
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - dTdtSOLAR(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- shortwave solar temperature tendency
- units :
- Celcius second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - dTdtRESTORE(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- restoring temperature tendency
- units :
- Celcius second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - dTdtFORCE(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- forcing temperature tendency
- units :
- Celcius second-1
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - dUdtRESTORE(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- restoring zonal velocity tendency
- units :
- meters second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - dUdtFORCE(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- forcing zonal velocity tendency
- units :
- meters second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - dVdtRESTORE(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- restoring meridional velocity tendency
- units :
- meters second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - dVdtFORCE(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- forcing merid velocity tendency
- units :
- meters second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - epsilon(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- subgrid-scale dissipation of kinetic energy
- units :
- meters2 second-3
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - N2(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- Brunt-Vaisalla frequency squared
- units :
- second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - S2(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- dudz2 + dvdz2
- units :
- second-2
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - RIG(longitude, month, time, z)float32dask.array<chunksize=(1, 1, 1313, 288), meta=np.ndarray>
- long_name :
- gradient Richardson number, of mean profile
- time :
- time
Array Chunk Bytes 1.44 MiB 1.44 MiB Shape (1, 1, 1313, 288) (1, 1, 1313, 288) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - nududztop(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. visc. flux u-momentum, top bry
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - nudvdztop(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. visc. flux v-momentum, top bry
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - kappadsdztop(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. diff. flux salt, top bry
- units :
- meter second-1 psu
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - kappadtdztop(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. diff. flux temp, top bry
- units :
- meter second-1 Celcius
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - nududzbot(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. visc. flux u-momentum, bot bry
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - nudvdzbot(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. visc. flux v-momentum, bot bry
- units :
- meter2 second-2
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - kappadsdzbot(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. diff. flux salt, bot bry
- units :
- meter second-1 psu
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - kappadtdzbot(longitude, month, time)float32dask.array<chunksize=(1, 1, 1313), meta=np.ndarray>
- long_name :
- vert. diff. flux temp, bot bry
- units :
- meter second-1 Celcius
- time :
- time
Array Chunk Bytes 5.13 kiB 5.13 kiB Shape (1, 1, 1313) (1, 1, 1313) Count 4 Tasks 1 Chunks Type float32 numpy.ndarray - alpha(longitude, month)float640.0002976
- long_name :
- thermal expansion coeff., lin. eq. of state
array([[0.00029755]])
- beta(longitude, month)float64-0.0007386
- long_name :
- haline contraction coeff., lin. eq. of state
array([[-0.00073863]])
- T0(longitude, month)float6425.0
- long_name :
- reference temperature, lin eq. of state
array([[25.]])
- S0(longitude, month)float6435.25
- long_name :
- reference salinity, lin. eq. of state
array([[35.25]])
- rho0(longitude, month)float641.024e+03
- long_name :
- reference density, lin. eq. of state
array([[1023.53]])
- type :
- DIABLO LES, processed means
- title :
- ROMS_PSH_6HRLIN_0N140W_360x360x288_5OCT2021
- history :
- Tue Dec 28 09:09:16 2021: ncatted -O -a units,time,m,c,seconds since 1985-05-25 03:00:00 ROMS_PSH_3HRLIN_242_602_360x360x288_25MAY1985_24NOV2021.nc
- NCO :
- netCDF Operators version 4.9.5 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
- intake_esm_varname :
- ('ume', 'vme', 'tempme', 'saltme', 'urms', 'vrms', 'wrms', 'temprms', 'saltrms', 'uw', 'vw', 'saltw', 'tempw', 'nududz', 'nudvdz', 'kappadsdz', 'kappadtdz', 'dTdtSOLAR', 'dTdtRESTORE', 'dTdtFORCE', 'dUdtRESTORE', 'dUdtFORCE', 'dVdtRESTORE', 'dVdtFORCE', 'epsilon', 'N2', 'S2', 'RIG', 'nududztop', 'nudvdztop', 'kappadsdztop', 'kappadtdztop', 'nududzbot', 'nudvdzbot', 'kappadsdzbot', 'kappadtdzbot', 'alpha', 'beta', 'T0', 'S0', 'rho0')
- intake_esm_dataset_key :
- -140.may